The molecular basis for centromere identity and function
- PMID: 26601620
- PMCID: PMC8603311
- DOI: 10.1038/nrm.2015.5
The molecular basis for centromere identity and function
Abstract
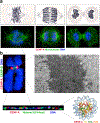
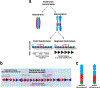
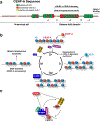
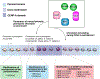
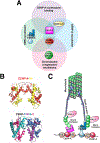
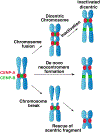
The centromere is the region of the chromosome that directs its segregation in mitosis and meiosis. Although the functional importance of the centromere has been appreciated for more than 130 years, elucidating the molecular features and properties that enable centromeres to orchestrate chromosome segregation is an ongoing challenge. Most eukaryotic centromeres are defined epigenetically and require the presence of nucleosomes containing the histone H3 variant centromere protein A (CENP-A; also known as CENH3). Ongoing work is providing important molecular insights into the central requirements for centromere identity and propagation, and the mechanisms by which centromeres recruit kinetochores to connect to spindle microtubules.
Figures
References
-
- Flemming W Zellsubstanz, Kern und Zelltheilung (Vogel, Leipzig, 1882).
-
- Bernard P et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–42 (2001). - PubMed
-
- Nasmyth K Segregating sister genomes: the molecular biology of chromosome separation. Science 297, 559–65 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

