The Tomato Nucleotide-binding Leucine-rich Repeat Immune Receptor I-2 Couples DNA-binding to Nucleotide-binding Domain Nucleotide Exchange
- PMID: 26601946
- PMCID: PMC4714197
- DOI: 10.1074/jbc.M115.698589
The Tomato Nucleotide-binding Leucine-rich Repeat Immune Receptor I-2 Couples DNA-binding to Nucleotide-binding Domain Nucleotide Exchange
Abstract
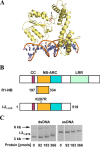
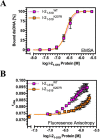
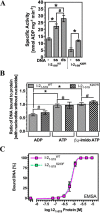
Plant nucleotide-binding leucine-rich repeat (NLR) proteins enable plants to recognize and respond to pathogen attack. Previously, we demonstrated that the Rx1 NLR of potato is able to bind and bend DNA in vitro. DNA binding in situ requires its genuine activation following pathogen perception. However, it is unknown whether other NLR proteins are also able to bind DNA. Nor is it known how DNA binding relates to the ATPase activity intrinsic to NLR switch function required to immune activation. Here we investigate these issues using a recombinant protein corresponding to the N-terminal coiled-coil and nucleotide-binding domain regions of the I-2 NLR of tomato. Wild type I-2 protein bound nucleic acids with a preference of ssDNA ≈ dsDNA > ssRNA, which is distinct from Rx1. I-2 induced bending and melting of DNA. Notably, ATP enhanced DNA binding relative to ADP in the wild type protein, the null P-loop mutant K207R, and the autoactive mutant S233F. DNA binding was found to activate the intrinsic ATPase activity of I-2. Because DNA binding by I-2 was decreased in the presence of ADP when compared with ATP, a cyclic mechanism emerges; activated ATP-associated I-2 binds to DNA, which enhances ATP hydrolysis, releasing ADP-bound I-2 from the DNA. Thus DNA binding is a general property of at least a subset of NLR proteins, and NLR activation is directly linked to its activity at DNA.
Keywords: ATPases associated with diverse cellular activities (AAA); DNA binding protein; Nod-like receptor (NLR); cellular immune response; nucleotide; plant biochemistry.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Dangl J. L., and Jones J. D. (2001) Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 - PubMed
-
- Jones J. D., and Dangl J. L. (2006) The plant immune system. Nature 444, 323–329 - PubMed
-
- Maekawa T., Kufer T. A., and Schulze-Lefert P. (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12, 817–826 - PubMed
-
- Medzhitov R. (2007) Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

