Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation
- PMID: 26603296
- PMCID: PMC4670565
- DOI: 10.1016/j.cmet.2015.09.025
Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation
Abstract
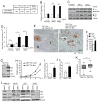
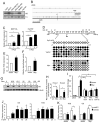
c-Myc is known to promote glutamine usage by upregulating glutaminase (GLS), which converts glutamine to glutamate that is catabolized in the TCA cycle. Here we report that in a number of human and murine cells and cancers, Myc induces elevated expression of glutamate-ammonia ligase (GLUL), also termed glutamine synthetase (GS), which catalyzes the de novo synthesis of glutamine from glutamate and ammonia. This is through upregulation of a Myc transcriptional target thymine DNA glycosylase (TDG), which promotes active demethylation of the GS promoter and its increased expression. Elevated expression of GS promotes cell survival under glutamine limitation, while silencing of GS decreases cell proliferation and xenograft tumor growth. Upon GS overexpression, increased glutamine enhances nucleotide synthesis and amino acid transport. These results demonstrate an unexpected role of Myc in inducing glutamine synthesis and suggest a molecular connection between DNA demethylation and glutamine metabolism in Myc-driven cancers.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


References
-
- Biolo G, Zorat F, Antonione R, Ciocchi B. Muscle glutamine depletion in the intensive care unit. The international journal of biochemistry & cell biology. 2005;37:2169–2179. - PubMed
-
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK053307/DK/NIDDK NIH HHS/United States
- R37 DK060596/DK/NIDDK NIH HHS/United States
- R01 CA163591/CA/NCI NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- U01 CA168409/CA/NCI NIH HHS/United States
- R01CA129536/CA/NCI NIH HHS/United States
- R01 CA129536/CA/NCI NIH HHS/United States
- R01GM97355/GM/NIGMS NIH HHS/United States
- P30 CA072720/CA/NCI NIH HHS/United States
- R37DK060596/DK/NIDDK NIH HHS/United States
- R01 GM097355/GM/NIGMS NIH HHS/United States
- R01 CA169919/CA/NCI NIH HHS/United States
- R01DK053307/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

