LPS impairs oxygen utilization in epithelia by triggering degradation of the mitochondrial enzyme Alcat1
- PMID: 26604221
- PMCID: PMC4732295
- DOI: 10.1242/jcs.176701
LPS impairs oxygen utilization in epithelia by triggering degradation of the mitochondrial enzyme Alcat1
Abstract
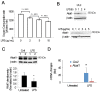
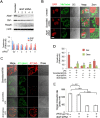
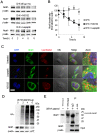
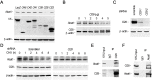
Cardiolipin (also known as PDL6) is an indispensable lipid required for mitochondrial respiration that is generated through de novo synthesis and remodeling. Here, the cardiolipin remodeling enzyme, acyl-CoA:lysocardiolipin-acyltransferase-1 (Alcat1; SwissProt ID, Q6UWP7) is destabilized in epithelia by lipopolysaccharide (LPS) impairing mitochondrial function. Exposure to LPS selectively decreased levels of carbon 20 (C20)-containing cardiolipin molecular species, whereas the content of C18 or C16 species was not significantly altered, consistent with decreased levels of Alcat1. Alcat1 is a labile protein that is lysosomally degraded by the ubiquitin E3 ligase Skp-Cullin-F-box containing the Fbxo28 subunit (SCF-Fbxo28) that targets Alcat1 for monoubiquitylation at residue K183. Interestingly, K183 is also an acetylation-acceptor site, and acetylation conferred stability to the enzyme. Histone deacetylase 2 (HDAC2) interacted with Alcat1, and expression of a plasmid encoding HDAC2 or treatment of cells with LPS deacetylated and destabilized Alcat1, whereas treatment of cells with a pan-HDAC inhibitor increased Alcat1 levels. Alcat1 degradation was partially abrogated in LPS-treated cells that had been silenced for HDAC2 or treated with MLN4924, an inhibitor of Cullin-RING E3 ubiquitin ligases. Thus, LPS increases HDAC2-mediated Alcat1 deacetylation and facilitates SCF-Fbxo28-mediated disposal of Alcat1, thus impairing mitochondrial integrity.
Keywords: Degradation; Mitochondria; Ubiquitin.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures




References
-
- Au P. Y., Argiropoulos B., Parboosingh J. S. and Micheil Innes A. (2014). Refinement of the critical region of 1q41q42 microdeletion syndrome identifies FBXO28 as a candidate causative gene for intellectual disability and seizures. Am. J. Med. Genet. A 164A, 441-448. 10.1002/ajmg.a.36320 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA165065/CA/NCI NIH HHS/United States
- HL01916/HL/NHLBI NIH HHS/United States
- R01 OH008282/OH/NIOSH CDC HHS/United States
- R01 HL112791/HL/NHLBI NIH HHS/United States
- R01 HL097376/HL/NHLBI NIH HHS/United States
- R01 HL116472/HL/NHLBI NIH HHS/United States
- P30 DK072506/DK/NIDDK NIH HHS/United States
- HL116472/HL/NHLBI NIH HHS/United States
- ES020693/ES/NIEHS NIH HHS/United States
- HL125435/HL/NHLBI NIH HHS/United States
- R01 HL125435/HL/NHLBI NIH HHS/United States
- P01 HL114453/HL/NHLBI NIH HHS/United States
- HL098174/HL/NHLBI NIH HHS/United States
- R01 HL081784/HL/NHLBI NIH HHS/United States
- CA 165065/CA/NCI NIH HHS/United States
- I01 BX002200/BX/BLRD VA/United States
- R01 ES020693/ES/NIEHS NIH HHS/United States
- HL097376/HL/NHLBI NIH HHS/United States
- U19AIO68021/PHS HHS/United States
- HL096376/HL/NHLBI NIH HHS/United States
- HL112791/HL/NHLBI NIH HHS/United States
- UH2 HL123502/HL/NHLBI NIH HHS/United States
- OH008282/OH/NIOSH CDC HHS/United States
- 1UH2HL123502/HL/NHLBI NIH HHS/United States
- HL081784/HL/NHLBI NIH HHS/United States
- P01HL114453/HL/NHLBI NIH HHS/United States
- R01 HL096376/HL/NHLBI NIH HHS/United States
- R01 HL086884/HL/NHLBI NIH HHS/United States
- R01 HL098174/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

