CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity
- PMID: 26607793
- PMCID: PMC4671820
- DOI: 10.1016/j.cell.2015.10.068
CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity
Abstract
Th17 cells play a critical role in host defense against extracellular pathogens and tissue homeostasis but can induce autoimmunity. The mechanisms implicated in balancing "pathogenic" and "non-pathogenic" Th17 cell states remain largely unknown. We used single-cell RNA-seq to identify CD5L/AIM as a regulator expressed in non-pathogenic, but not in pathogenic Th17 cells. Although CD5L does not affect Th17 differentiation, it is a functional switch that regulates the pathogenicity of Th17 cells. Loss of CD5L converts non-pathogenic Th17 cells into pathogenic cells that induce autoimmunity. CD5L mediates this effect by modulating the intracellular lipidome, altering fatty acid composition and restricting cholesterol biosynthesis and, thus, ligand availability for Rorγt, the master transcription factor of Th17 cells. Our study identifies CD5L as a critical regulator of the Th17 cell functional state and highlights the importance of lipid metabolism in balancing immune protection and disease induced by T cells.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
During the course of the research, VKK had an ownership interest in Tempero Pharmaceuticals, a company that was working in the area of Treg-Th17 biology and developing treatments for autoimmune diseases in areas related to this research. VK’s ownership ended in October 2014. VK’s interests were reviewed and managed by the Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Figures
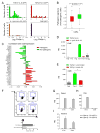


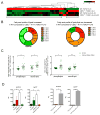

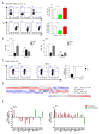
Comment in
-
T cells: Seq-ing out the 'bad' guys.Nat Rev Immunol. 2016 Jan;16(1):3. doi: 10.1038/nri.2015.15. Epub 2015 Dec 21. Nat Rev Immunol. 2016. PMID: 26688347 No abstract available.
References
-
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nature genetics. 2007;39:503–512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI039671/AI/NIAID NIH HHS/United States
- P01 AI056299/AI/NIAID NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- P01 NS076410/NS/NINDS NIH HHS/United States
- P01 AI039671/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P01 AI045757/AI/NIAID NIH HHS/United States
- 4R00AI110649/AI/NIAID NIH HHS/United States
- AI056299/AI/NIAID NIH HHS/United States
- R29 NS030843/NS/NINDS NIH HHS/United States
- NS076410/NS/NINDS NIH HHS/United States
- R01 NS030843/NS/NINDS NIH HHS/United States
- R00 AI110649/AI/NIAID NIH HHS/United States
- NS030843/NS/NINDS NIH HHS/United States
- R37 NS030843/NS/NINDS NIH HHS/United States
- P50 HG006193/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

