High-Throughput Single-Cell Kinetics of Virus Infections in the Presence of Defective Interfering Particles
- PMID: 26608322
- PMCID: PMC4719634
- DOI: 10.1128/JVI.02190-15
High-Throughput Single-Cell Kinetics of Virus Infections in the Presence of Defective Interfering Particles
Abstract
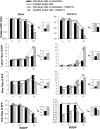
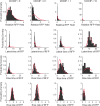
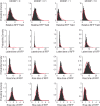
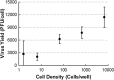
Defective interfering particles (DIPs) are virus mutants that lack essential genes for growth. In coinfections with helper virus, the diversion of viral proteins to the replication and packaging of DIP genomes can interfere with virus production. Mounting cases of DIPs and DIP-like genomes in clinical and natural isolates, as well as growing interest in DIP-based therapies, underscore a need to better elucidate how DIPs work. DIP activity is primarily measured by its inhibition of virus infection yield, an endpoint that masks the dynamic and potentially diverse individual cell behaviors. Using vesicular stomatitis virus (VSV) as a model, we coinfected BHK cells with VSV DIPs and recombinant helper virus carrying a gene encoding a red fluorescent protein (RFP) whose expression correlates with the timing and level of virus release. For single cells within a monolayer, 10 DIPs per cell suppressed the reporter expression in only 1.2% of the cells. In most cells, it slowed and reduced viral gene expression, manifested as a shift in mean latent time from 4 to 6 h and reduced virus yields by 10-fold. For single cells isolated in microwells, DIP effects were more pronounced, reducing virus yields by 100-fold and extending latent times to 12 h, including individual instances above 20 h. Together, these results suggest that direct or indirect cell-cell interactions prevent most coinfected cells from being completely suppressed by DIPs. Finally, a gamma distribution model captures well how the infection kinetics quantitatively depends on the DIP dose. Such models will be useful for advancing a predictive biology of DIP-associated virus growth and infection spread.
Importance: During the last century, basic studies in virology have focused on developing a molecular mechanistic understanding of how infectious viruses reproduce in their living host cells. However, over the last 10 years, the advent of deep sequencing and other powerful technologies has revealed in natural and patient infections that viruses do not act alone. Instead, viruses are often accompanied by defective virus-like particles that carry large deletions in their genomes and fail to replicate on their own. Coinfections of viable and defective viruses behave in unpredictable ways, but they often interfere with normal virus growth, potentially enabling infections to evade host immune surveillance. In the current study, controlled levels of defective viruses are coinfected with viable viruses that have been engineered to express a fluorescent reporter protein during infection. Unique profiles of reporter expression acquired from thousands of coinfected cells reveal how interference acts at multiple stages of infection.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

