RTTN Mutations Cause Primary Microcephaly and Primordial Dwarfism in Humans
- PMID: 26608784
- PMCID: PMC4678428
- DOI: 10.1016/j.ajhg.2015.10.012
RTTN Mutations Cause Primary Microcephaly and Primordial Dwarfism in Humans
Abstract
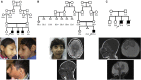
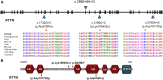
Primary microcephaly is a developmental brain anomaly that results from defective proliferation of neuroprogenitors in the germinal periventricular zone. More than a dozen genes are known to be mutated in autosomal-recessive primary microcephaly in isolation or in association with a more generalized growth deficiency (microcephalic primordial dwarfism), but the genetic heterogeneity is probably more extensive. In a research protocol involving autozygome mapping and exome sequencing, we recruited a multiplex consanguineous family who is affected by severe microcephalic primordial dwarfism and tested negative on clinical exome sequencing. Two candidate autozygous intervals were identified, and the second round of exome sequencing revealed a single intronic variant therein (c.2885+8A>G [p.Ser963(∗)] in RTTN exon 23). RT-PCR confirmed that this change creates a cryptic splice donor and thus causes retention of the intervening 7 bp of the intron and leads to premature truncation. On the basis of this finding, we reanalyzed the exome file of a second consanguineous family affected by a similar phenotype and identified another homozygous change in RTTN as the likely causal mutation. Combined linkage analysis of the two families confirmed that RTTN maps to the only significant linkage peak. Finally, through international collaboration, a Canadian multiplex family affected by microcephalic primordial dwarfism and biallelic mutation of RTTN was identified. Our results expand the phenotype of RTTN-related disorders, hitherto limited to polymicrogyria, to include microcephalic primordial dwarfism with a complex brain phenotype involving simplified gyration.
Keywords: ciliopathy; lissencephaly; primary cilium; primordial dwarfism; rotatin.
Copyright © 2015 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

