De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly
- PMID: 26609813
- PMCID: PMC4709270
- DOI: 10.7554/eLife.10586
De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly
Abstract
Vertebrate centrioles normally propagate through duplication, but in the absence of preexisting centrioles, de novo synthesis can occur. Consistently, centriole formation is thought to strictly rely on self-assembly, involving self-oligomerization of the centriolar protein SAS-6. Here, through reconstitution of de novo synthesis in human cells, we surprisingly found that normal looking centrioles capable of duplication and ciliation can arise in the absence of SAS-6 self-oligomerization. Moreover, whereas canonically duplicated centrioles always form correctly, de novo centrioles are prone to structural errors, even in the presence of SAS-6 self-oligomerization. These results indicate that centriole biogenesis does not strictly depend on SAS-6 self-assembly, and may require preexisting centrioles to ensure structural accuracy, fundamentally deviating from the current paradigm.
Keywords: SAS-6; cartwheel; cell biology; centriole; centrosome; cilia; human; self-assembly.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
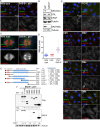


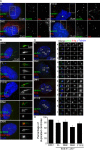
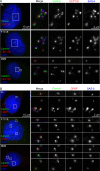

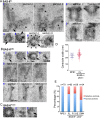
References
-
- Abumuslimov SS, Nadezhdina ES, Chentsov I. An electron microscopic study of centriole and centrosome morphogenesis in the early development of the mouse. Tsitologiia. 1994;36:1054–1061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

