Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family
- PMID: 26610890
- PMCID: PMC4767052
- DOI: 10.1093/glycob/cwv108
Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family
Abstract
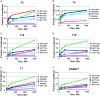
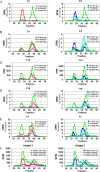
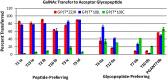
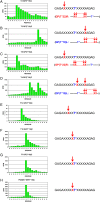
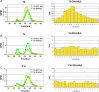
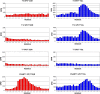
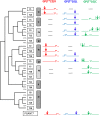
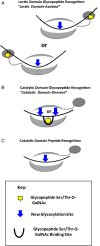
A large family of UDP-GalNAc:polypeptide GalNAc transferases (ppGalNAc-Ts) initiates and defines sites of mucin-type Ser/Thr-O-GalNAc glycosylation. Family members have been classified into peptide- and glycopeptide-preferring subfamilies, although both families possess variable activities against glycopeptide substrates. All but one isoform contains a C-terminal carbohydrate-binding lectin domain whose roles in modulating glycopeptide specificity is just being understood. We have previously shown for several peptide-preferring isoforms that the presence of a remote Thr-O-GalNAc, 6-17 residues from a Ser/Thr acceptor site, may enhance overall catalytic activity in an N- or C-terminal direction. This enhancement varies with isoform and is attributed to Thr-O-GalNAc interactions at the lectin domain. We now report on the glycopeptide substrate utilization of a series of glycopeptide (human-ppGalNAc-T4, T7, T10, T12 and fly PGANT7) and peptide-preferring transferases (T2, T3 and T5) by exploiting a series of random glycopeptide substrates designed to probe the functions of their catalytic and lectin domains. Glycosylation was observed at the -3, -1 and +1 residues relative to a neighboring Thr-O-GalNAc, depending on isoform, which we attribute to specific Thr-O-GalNAc binding at the catalytic domain. Additionally, these glycopeptide-preferring isoforms show remote lectin domain-assisted Thr-O-GalNAc enhancements that vary from modest to none. We conclude that the glycopeptide specificity of the glycopeptide-preferring isoforms predominantly resides in their catalytic domain but may be further modulated by remote lectin domain interactions. These studies further demonstrate that both domains of the ppGalNAc-Ts have specialized and unique functions that work in concert to control and order mucin-type O-glycosylation.
Keywords: glycoprotein biosynthesis; glycosyltransferase; lectin; mucin.
© The Author 2015. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Bennett EP, Hassan H, Hollingsworth MA, Clausen H. 1999. A novel human UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 460:226–230. - PubMed
-
- Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG et al. . 1998. Cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem. 273:30472–30481. - PubMed
-
- Berois N, Mazal D, Ubillos L, Trajtenberg F, Nicolas A, Sastre-Garau X, Magdelenat H, Osinaga E. 2006. UDP-N-acetyl-D-galactosamine: Polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. J Histochem Cytochem. 54:317–328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

