Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus
- PMID: 26611651
- PMCID: PMC7114344
- DOI: 10.1016/j.prevetmed.2015.10.020
Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus
Abstract
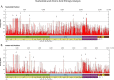
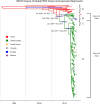
Porcine epidemic diarrhea virus (PEDV) has caused severe economic losses both recently in the United States (US) and historically throughout Europe and Asia. Traditionally, analysis of the spike gene has been used to determine phylogenetic relationships between PEDV strains. We determined the complete genomes of 93 PEDV field samples from US swine and analyzed the data in conjunction with complete genome sequences available from GenBank (n=126) to determine the most variable genomic areas. Our results indicate high levels of variation within the ORF1 and spike regions while the C-terminal domains of structural genes were highly conserved. Analysis of the Receptor Binding Domains in the spike gene revealed a limited number of amino acid substitutions in US strains compared to Asian strains. Phylogenetic analysis of the complete genome sequence data revealed high rates of recombination, resulting in differing evolutionary patterns in phylogenies inferred for the spike region versus whole genomes. These finding suggest that significant genetic events outside of the spike region have contributed to the evolution of PEDV.
Keywords: Bayesian analysis; Complete genome; Molecular analysis; Porcine epidemic diarrhea virus.
Copyright © 2015 The Authors. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52:234–243. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

