Functional characterization of a human POU1F1 mutation associated with isolated growth hormone deficiency: a novel etiology for IGHD
- PMID: 26612202
- PMCID: PMC5007599
- DOI: 10.1093/hmg/ddv486
Functional characterization of a human POU1F1 mutation associated with isolated growth hormone deficiency: a novel etiology for IGHD
Abstract
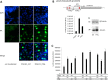
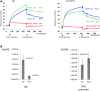
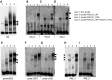
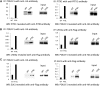
POU1F1, a pituitary-specific POU-homeo domain transcription factor, plays an essential role in the specification of the somatotroph, lactotroph and thyrotroph lineages and in the activation of GH1, PRL and TSHβ transcription. Individuals with mutations in POU1F1 present with combined deficiency of GH, PRL and TSH. Here, we identified a heterozygous missense mutation with evidence of pathogenicity, at the POU1F1 locus, in a large family in which an isolated growth hormone deficiency segregates as an autosomal dominant trait. The corresponding p.Pro76Leu mutation maps to a conserved site within the POU1F1 transactivation domain. Bandshift assays revealed that the mutation alters wild-type POU1F1 binding to cognate sites within the hGH-LCR and hGH1 promoter, but not to sites within the PRL promoter, and it selectively increases binding affinity to sites within the hGH-LCR. Co-immunoprecipitation studies reveal that this substitution enhances interactions of POU1F1 with three of its cofactors, PITX1, LHX3a and ELK1, and that residue 76 plays a critical role in these interactions. The insertion of the mutation at the mouse Pou1f1 locus results in a dramatic loss of protein expression despite normal mRNA concentrations. Mice heterozygous for the p.Pro76Leu mutation were phenotypically normal while homozygotes demonstrated a dwarf phenotype. Overall, this study unveils the involvement of POU1F1 in dominantly inherited isolated GH deficiency and demonstrates a significant impact of the Pro76Leu mutation on DNA-binding activities, alterations in transactivating functions and interactions with cofactors. Our data further highlight difficulties in modeling human genetic disorders in the mouse despite apparent conservation of gene expression pathways and physiologic functions.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Andersen B., Rosenfeld M.G. (2001) POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev., 22, 2–35. - PubMed
-
- Ingraham H.A., Flynn S.E., Voss J.W., Albert V.R., Kapiloff M.S., Wilson L., Rosenfeld M.G. (1990) The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1-Pit-1 interactions. Cell, 61, 1021–1033. - PubMed
-
- Andersen B., Rosenfeld M.G. (1994) Pit-1 determines cell types during development of the anterior pituitary gland. A model for transcriptional regulation of cell phenotypes in mammalian organogenesis. J. Biol. Chem., 269, 29335–29338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

