Uncovering diversity in the development of central noradrenergic neurons and their efferents
- PMID: 26612521
- PMCID: PMC4901874
- DOI: 10.1016/j.brainres.2015.11.023
Uncovering diversity in the development of central noradrenergic neurons and their efferents
Abstract
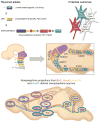
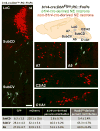
Uncovering the mechanisms that underlie central noradrenergic neuron heterogeneity is essential to understanding selective subtype vulnerability to disease and environmental insult. Using recombinase-based intersectional genetic fate mapping we have previously demonstrated that molecularly distinct progenitor populations give rise to mature noradrenergic neurons differing in their anatomical location, axon morphology and efferent projection pattern. Here we review the findings from our previous study and extend our analysis of the noradrenergic subpopulation defined by transient developmental expression of Hoxb1. Using a combination of intersectional genetic fate mapping and analysis of a targeted loss of function mutation in Hoxb1, we have now uncovered additional heterogeneity based on the requirement of some noradrenergic neurons for Hoxb1 expression. By comparing the distribution of noradrenergic neurons derived from the Hoxb1 expression domain in wild-type and mutant mice, we demonstrate that Hoxb1 expression is required by a subset of neurons in the pons. Additional fate mapping, using a Hoxb1 enhancer element that drives Cre recombinase expression exclusively in rhombomere 4 of the hindbrain, reveals the existence of a subpopulation of noradrenergic neurons in the pons with more restricted axonal targets than the full Hoxb1-derived subpopulation. The unique projection profile of this newly defined subpopulation suggests that it may be functionally distinct. These analyses shed new light on the molecular determinants of noradrenergic identity in the pons and the overall complexity of the central noradrenergic system. This article is part of a Special Issue entitled SI: Noradrenergic System.
Keywords: Efferents; Fate map; Hoxb1; Norepinephrine; Recombinase-based intersectional genetic fate mapping; Rhombomere.
Published by Elsevier B.V.
Figures




References
-
- Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nature genetics. 2003;35:70–5. - PubMed
-
- Berridge CW, Waterhouse BD. The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain research. Brain research reviews. 2003;42:33–84. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

