DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts
- PMID: 26612864
- PMCID: PMC4702897
- DOI: 10.1093/nar/gkv1270
DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts
Abstract
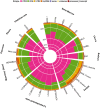
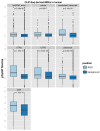
microRNAs (miRNAs) are short non-coding RNAs (ncRNAs) that act as post-transcriptional regulators of coding gene expression. Long non-coding RNAs (lncRNAs) have been recently reported to interact with miRNAs. The sponge-like function of lncRNAs introduces an extra layer of complexity in the miRNA interactome. DIANA-LncBase v1 provided a database of experimentally supported and in silico predicted miRNA Recognition Elements (MREs) on lncRNAs. The second version of LncBase (www.microrna.gr/LncBase) presents an extensive collection of miRNA:lncRNA interactions. The significantly enhanced database includes more than 70 000 low and high-throughput, (in)direct miRNA:lncRNA experimentally supported interactions, derived from manually curated publications and the analysis of 153 AGO CLIP-Seq libraries. The new experimental module presents a 14-fold increase compared to the previous release. LncBase v2 hosts in silico predicted miRNA targets on lncRNAs, identified with the DIANA-microT algorithm. The relevant module provides millions of predicted miRNA binding sites, accompanied with detailed metadata and MRE conservation metrics. LncBase v2 caters information regarding cell type specific miRNA:lncRNA regulation and enables users to easily identify interactions in 66 different cell types, spanning 36 tissues for human and mouse. Database entries are also supported by accurate lncRNA expression information, derived from the analysis of more than 6 billion RNA-Seq reads.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

