The autoinhibitory CARD2-Hel2i Interface of RIG-I governs RNA selection
- PMID: 26612866
- PMCID: PMC4737149
- DOI: 10.1093/nar/gkv1299
The autoinhibitory CARD2-Hel2i Interface of RIG-I governs RNA selection
Abstract
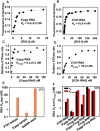
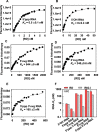
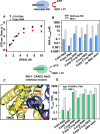
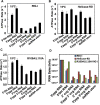
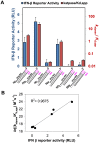
RIG-I (Retinoic Acid Inducible Gene-I) is a cytosolic innate immune receptor that detects atypical features in viral RNAs as foreign to initiate a Type I interferon signaling response. RIG-I is present in an autoinhibited state in the cytoplasm and activated by blunt-ended double-stranded (ds)RNAs carrying a 5' triphosphate (ppp) moiety. These features found in many pathogenic RNAs are absent in cellular RNAs due to post-transcriptional modifications of RNA ends. Although RIG-I is structurally well characterized, the mechanistic basis for RIG-I's remarkable ability to discriminate between cellular and pathogenic RNAs is not completely understood. We show that RIG-I's selectivity for blunt-ended 5'-ppp dsRNAs is ≈3000 times higher than non-blunt ended dsRNAs commonly found in cellular RNAs. Discrimination occurs at multiple stages and signaling RNAs have high affinity and ATPase turnover rate and thus a high katpase/Kd. We show that RIG-I uses its autoinhibitory CARD2-Hel2i (second CARD-helicase insertion domain) interface as a barrier to select against non-blunt ended dsRNAs. Accordingly, deletion of CARDs or point mutations in the CARD2-Hel2i interface decreases the selectivity from ≈3000 to 150 and 750, respectively. We propose that the CARD2-Hel2i interface is a 'gate' that prevents cellular RNAs from generating productive complexes that can signal.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M., Jr, Akira S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. - PubMed
-
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

