Phospholipase C Beta 1: a Candidate Signature Gene for Proneural Subtype High-Grade Glioma
- PMID: 26614510
- PMCID: PMC5085994
- DOI: 10.1007/s12035-015-9518-2
Phospholipase C Beta 1: a Candidate Signature Gene for Proneural Subtype High-Grade Glioma
Abstract
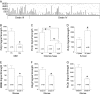
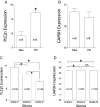
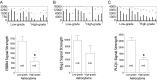
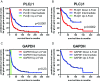
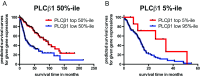
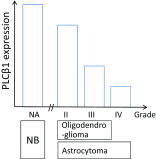
Phospholipase C beta 1 (PLCβ1) expresses in gliomas and cultured glial cells, but its expression is barely detectable in normal glial cells. We analyzed data from Gene Expression Omnibus (GEO-GDSxxx), The Cancer Genome Atlas (TCGA), and the Repository for Molecular Brain Neoplasia Data (REMBRANDT) to explore the potential role of PLCβ1 as a biomarker in high-grade glioma (HGG). PLCβ1 expression is significantly higher in grade III gliomas than that in grade IV gliomas from GDS1815 (n = 24 vs. 76), GDS1962 (n = 19 vs. 81), and GDS1975 (n = 26 vs. 59). In GDS1815, PLCβ1 expression correlates with several known proneural (PN) signature genes; its expression from PN subtype (n = 15) is significantly higher than that from mesenchymal (Mes) subtype (n = 33) HGG. In GDS1962, PLCβ1 expression is the highest in nontumor brain tissue (n = 23) and is significantly higher than its expression in grade II gliomas [astrocytomas (n = 7) and oligodendrogliomas (n = 37)]. A Kaplan-Meier survival curve from a REMBRANDT cohort demonstrates that glioma patients with intermediate PLCβ1 expression (n = 103) survived significantly longer than PLCβ1 downregulated (2X) groups (n = 226). From TCGA data, PLCβ1 RNA-Seq signal inversely correlates with the pathological grades, and PLCβ1 expression in PN (n = 8) is of significantly higher levels than that in Mes (n = 8) subtypes of glioblastoma. The top 50 % of PLCβ1 expression subgroup (n = 294) of gliomas (grades II to IV merged) survived significantly longer than the low 50 percentile of the PLCβ1 expression subgroup (n = 293). p values are less than 0.05 for all these analyses. We conclude that PLCβ1 is a candidate signature gene for PN subtype HGG, and its expression inversely correlates with glioma pathological grade and is a potential prognostic factor.
Keywords: Biomarker; Glioblastoma; Glioma; PLCβ1; Proneural; REMBRANDT; Signature gene; TCGA.
Conflict of interest statement
Compliance with Ethical Standards Funding J.-J.Z. and M.L. received funding support from Dr. Marnie Rose Foundation and J.T.C from NIH TL1TR000371. Conflict of Interest None.
Figures


Similar articles
-
Transforming growth factor beta induced (TGFBI) is a potential signature gene for mesenchymal subtype high-grade glioma.J Neurooncol. 2018 Apr;137(2):395-407. doi: 10.1007/s11060-017-2729-9. Epub 2018 Jan 2. J Neurooncol. 2018. PMID: 29294230
-
The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas.PLoS One. 2010 Sep 3;5(9):e12548. doi: 10.1371/journal.pone.0012548. PLoS One. 2010. PMID: 20838435 Free PMC article.
-
Impact of phospholipase C β1 in glioblastoma: a study on the main mechanisms of tumor aggressiveness.Cell Mol Life Sci. 2022 Mar 18;79(4):195. doi: 10.1007/s00018-022-04198-1. Cell Mol Life Sci. 2022. PMID: 35303162 Free PMC article.
-
Isocitrate dehydrogenase status and molecular subclasses of glioma and glioblastoma.Neurosurg Focus. 2014 Dec;37(6):E13. doi: 10.3171/2014.9.FOCUS14505. Neurosurg Focus. 2014. PMID: 25434382 Review.
-
The Cancer Genome Atlas expression profiles of low-grade gliomas.Neurosurg Focus. 2014 Apr;36(4):E23. doi: 10.3171/2012.12.focus12351. Neurosurg Focus. 2014. PMID: 24812719 Review.
Cited by
-
Cryptic mutations of PLC family members in brain disorders: recent discoveries and a deep-learning-based approach.Brain. 2023 Apr 19;146(4):1267-1280. doi: 10.1093/brain/awac451. Brain. 2023. PMID: 36448305 Free PMC article. Review.
-
Large-scale analysis reveals a novel risk score to predict overall survival in hepatocellular carcinoma.Cancer Manag Res. 2018 Nov 21;10:6079-6096. doi: 10.2147/CMAR.S181396. eCollection 2018. Cancer Manag Res. 2018. PMID: 30538557 Free PMC article.
-
A novel gene signature based on five glioblastoma stem-like cell relevant genes predicts the survival of primary glioblastoma.J Cancer Res Clin Oncol. 2018 Mar;144(3):439-447. doi: 10.1007/s00432-017-2572-6. Epub 2018 Jan 3. J Cancer Res Clin Oncol. 2018. PMID: 29299749 Free PMC article.
-
The roles of phospholipase C-β related signals in the proliferation, metastasis and angiogenesis of malignant tumors, and the corresponding protective measures.Front Oncol. 2023 Jul 28;13:1231875. doi: 10.3389/fonc.2023.1231875. eCollection 2023. Front Oncol. 2023. PMID: 37576896 Free PMC article. Review.
-
Molecular regulation of PLCβ signaling.Methods Enzymol. 2023;682:17-52. doi: 10.1016/bs.mie.2023.01.001. Epub 2023 Feb 22. Methods Enzymol. 2023. PMID: 36948701 Free PMC article. Review.
References
-
- Okamoto Y, Di Patre PL, Burkhard C, Horstmann S, Jourde B, Fahey M, Schuler D, Probst-Hensch NM, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. - DOI - PubMed
-
- Figarella-Branger D, Labrousse F, Mohktari K, Societe Francaise de n. and Reseau de Neuro-Oncologie P Guidelines for adult diffuse gliomas WHO grade II, III and IV: pathology and biology. Societe francaise de neuropathologie. Reseau de neuro-oncologie pathologique. Ann Pathol. 2012;32:318–327. doi: 10.1016/j.annpat.2012.09.228. - DOI - PubMed
-
- Zinn PO, Colen RR, Kasper EM, Burkhardt JK. Extent of resection and radiotherapy in GBM: a 1973 to 2007 surveillance, epidemiology and end results analysis of 21,783 patients. Int J Oncol. 2013;42:929–934. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

