Centromere Locations in Brassica A and C Genomes Revealed Through Half-Tetrad Analysis
- PMID: 26614742
- PMCID: PMC4788232
- DOI: 10.1534/genetics.115.183210
Centromere Locations in Brassica A and C Genomes Revealed Through Half-Tetrad Analysis
Abstract
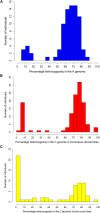
Locating centromeres on genome sequences can be challenging. The high density of repetitive elements in these regions makes sequence assembly problematic, especially when using short-read sequencing technologies. It can also be difficult to distinguish between active and recently extinct centromeres through sequence analysis. An effective solution is to identify genetically active centromeres (functional in meiosis) by half-tetrad analysis. This genetic approach involves detecting heterozygosity along chromosomes in segregating populations derived from gametes (half-tetrads). Unreduced gametes produced by first division restitution mechanisms comprise complete sets of nonsister chromatids. Along these chromatids, heterozygosity is maximal at the centromeres, and homologous recombination events result in homozygosity toward the telomeres. We genotyped populations of half-tetrad-derived individuals (from Brassica interspecific hybrids) using a high-density array of physically anchored SNP markers (Illumina Brassica 60K Infinium array). Mapping the distribution of heterozygosity in these half-tetrad individuals allowed the genetic mapping of all 19 centromeres of the Brassica A and C genomes to the reference Brassica napus genome. Gene and transposable element density across the B. napus genome were also assessed and corresponded well to previously reported genetic map positions. Known centromere-specific sequences were located in the reference genome, but mostly matched unanchored sequences, suggesting that the core centromeric regions may not yet be assembled into the pseudochromosomes of the reference genome. The increasing availability of genetic markers physically anchored to reference genomes greatly simplifies the genetic and physical mapping of centromeres using half-tetrad analysis. We discuss possible applications of this approach, including in species where half-tetrads are currently difficult to isolate.
Keywords: Brassica; centromeres; molecular karyotyping; recombination; unreduced gametes.
Copyright © 2016 by the Genetics Society of America.
Figures

References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Baudry E., Kryger P., Allsopp M., Koeniger N., Vautrin D., et al. , 2004. Whole-genome scan in thelytokous-laying workers of the cape honeybee (Apis mellifera capensis): central fusion, reduced recombination rates and centromere mapping using half-tetrad analysis. Genetics 167: 243–252. - PMC - PubMed
-
- Bretagnolle F., Thompson J. D., 1995. Tansley review No. 78. Gametes with the stomatic (sic) chromosome number: mechanisms of their formation and role in the evolution of autopolypoid plants. New Phytol. 129: 1–22. - PubMed
-
- Brownfield L., Köhler C., 2011. Unreduced gamete formation in plants: mechanisms and prospects. J. Exp. Bot. 62: 1659–1668. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

