Towards improved genome-scale metabolic network reconstructions: unification, transcript specificity and beyond
- PMID: 26615025
- PMCID: PMC5142010
- DOI: 10.1093/bib/bbv100
Towards improved genome-scale metabolic network reconstructions: unification, transcript specificity and beyond
Abstract
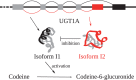
Genome-scale metabolic network reconstructions provide a basis for the investigation of the metabolic properties of an organism. There are reconstructions available for multiple organisms, from prokaryotes to higher organisms and methods for the analysis of a reconstruction. One example is the use of flux balance analysis to improve the yields of a target chemical, which has been applied successfully. However, comparison of results between existing reconstructions and models presents a challenge because of the heterogeneity of the available reconstructions, for example, of standards for presenting gene-protein-reaction associations, nomenclature of metabolites and reactions or selection of protonation states. The lack of comparability for gene identifiers or model-specific reactions without annotated evidence often leads to the creation of a new model from scratch, as data cannot be properly matched otherwise. In this contribution, we propose to improve the predictive power of metabolic models by switching from gene-protein-reaction associations to transcript-isoform-reaction associations, thus taking advantage of the improvement of precision in gene expression measurements. To achieve this precision, we discuss available databases that can be used to retrieve this type of information and point at issues that can arise from their neglect. Further, we stress issues that arise from non-standardized building pipelines, like inconsistencies in protonation states. In addition, problems arising from the use of non-specific cofactors, e.g. artificial futile cycles, are discussed, and finally efforts of the metabolic modelling community to unify model reconstructions are highlighted.
Keywords: gene-protein-reaction association; metabolic network reconstruction; unification.
© The Author 2015. Published by Oxford University Press.
Figures
Similar articles
-
Automated generation of genome-scale metabolic draft reconstructions based on KEGG.BMC Bioinformatics. 2018 Dec 4;19(1):467. doi: 10.1186/s12859-018-2472-z. BMC Bioinformatics. 2018. PMID: 30514205 Free PMC article.
-
Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis.PLoS Comput Biol. 2011 Mar;7(3):e1001116. doi: 10.1371/journal.pcbi.1001116. Epub 2011 Mar 31. PLoS Comput Biol. 2011. PMID: 21483480 Free PMC article.
-
Optimization based automated curation of metabolic reconstructions.BMC Bioinformatics. 2007 Jun 20;8:212. doi: 10.1186/1471-2105-8-212. BMC Bioinformatics. 2007. PMID: 17584497 Free PMC article.
-
Reconciliation of metabolites and biochemical reactions for metabolic networks.Brief Bioinform. 2014 Jan;15(1):123-35. doi: 10.1093/bib/bbs058. Epub 2012 Nov 19. Brief Bioinform. 2014. PMID: 23172809 Free PMC article. Review.
-
Physiologically Shrinking the Solution Space of a Saccharomyces cerevisiae Genome-Scale Model Suggests the Role of the Metabolic Network in Shaping Gene Expression Noise.PLoS One. 2015 Oct 8;10(10):e0139590. doi: 10.1371/journal.pone.0139590. eCollection 2015. PLoS One. 2015. PMID: 26448560 Free PMC article. Review.
Cited by
-
EColiCore2: a reference network model of the central metabolism of Escherichia coli and relationships to its genome-scale parent model.Sci Rep. 2017 Jan 3;7:39647. doi: 10.1038/srep39647. Sci Rep. 2017. PMID: 28045126 Free PMC article.
-
Constraint Based Modeling Going Multicellular.Front Mol Biosci. 2016 Feb 10;3:3. doi: 10.3389/fmolb.2016.00003. eCollection 2016. Front Mol Biosci. 2016. PMID: 26904548 Free PMC article. Review.
-
IDARE2-Simultaneous Visualisation of Multiomics Data in Cytoscape.Metabolites. 2021 May 6;11(5):300. doi: 10.3390/metabo11050300. Metabolites. 2021. PMID: 34066448 Free PMC article.
-
Assessing key decisions for transcriptomic data integration in biochemical networks.PLoS Comput Biol. 2019 Jul 19;15(7):e1007185. doi: 10.1371/journal.pcbi.1007185. eCollection 2019 Jul. PLoS Comput Biol. 2019. PMID: 31323017 Free PMC article.
-
Towards the network-based prediction of repurposed drugs using patient-specific metabolic models.EBioMedicine. 2019 May;43:26-27. doi: 10.1016/j.ebiom.2019.04.017. Epub 2019 Apr 9. EBioMedicine. 2019. PMID: 30979684 Free PMC article. No abstract available.
References
-
- Kim TY, Sohn SB, Kim YB, et al. Recent advances in reconstruction and applications of genome-scale metabolic models. Curr Opin Biotechnol 2012;23:617–23. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

