Genome-wide expression analysis offers new insights into the origin and evolution of Physcomitrella patens stress response
- PMID: 26615914
- PMCID: PMC4663497
- DOI: 10.1038/srep17434
Genome-wide expression analysis offers new insights into the origin and evolution of Physcomitrella patens stress response
Abstract
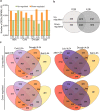
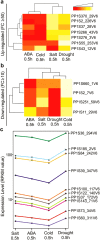
Changes in the environment, such as those caused by climate change, can exert stress on plant growth, diversity and ultimately global food security. Thus, focused efforts to fully understand plant response to stress are urgently needed in order to develop strategies to cope with the effects of climate change. Because Physcomitrella patens holds a key evolutionary position bridging the gap between green algae and higher plants, and because it exhibits a well-developed stress tolerance, it is an excellent model for such exploration. Here, we have used Physcomitrella patens to study genome-wide responses to abiotic stress through transcriptomic analysis by a high-throughput sequencing platform. We report a comprehensive analysis of transcriptome dynamics, defining profiles of elicited gene regulation responses to abiotic stress-associated hormone Abscisic Acid (ABA), cold, drought, and salt treatments. We identified more than 20,000 genes expressed under each aforementioned stress treatments, of which 9,668 display differential expression in response to stress. The comparison of Physcomitrella patens stress regulated genes with unicellular algae, vascular and flowering plants revealed genomic delineation concomitant with the evolutionary movement to land, including a general gene family complexity and loss of genes associated with different functional groups.
Figures





References
-
- Rensing S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008). - PubMed
-
- Floyd S. K. & Bowman J. L. The ancestral developmental tool kit of land plants. Int J Plant Sci 168, 1–35 (2007).
-
- Khraiwesh B. & Qudeimat E. in Climate change and plant abiotic Stress Tolerance 481-506 (Wiley-VCH Verlag GmbH & Co. KGaA, 2013).
-
- Knight H. & Knight M. R. Abiotic stress signalling pathways: specificity and cross-talk. Trends in plant science 6, 262–267 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

