The epigenomic landscape of African rainforest hunter-gatherers and farmers
- PMID: 26616214
- PMCID: PMC4674682
- DOI: 10.1038/ncomms10047
The epigenomic landscape of African rainforest hunter-gatherers and farmers
Abstract
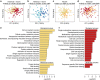
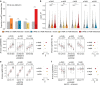
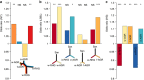
The genetic history of African populations is increasingly well documented, yet their patterns of epigenomic variation remain uncharacterized. Moreover, the relative impacts of DNA sequence variation and temporal changes in lifestyle and habitat on the human epigenome remain unknown. Here we generate genome-wide genotype and DNA methylation profiles for 362 rainforest hunter-gatherers and sedentary farmers. We find that the current habitat and historical lifestyle of a population have similarly critical impacts on the methylome, but the biological functions affected strongly differ. Specifically, methylation variation associated with recent changes in habitat mostly concerns immune and cellular functions, whereas that associated with historical lifestyle affects developmental processes. Furthermore, methylation variation--particularly that correlated with historical lifestyle--shows strong associations with nearby genetic variants that, moreover, are enriched in signals of natural selection. Our work provides new insight into the genetic and environmental factors affecting the epigenomic landscape of human populations over time.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

