Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci
- PMID: 26616563
- PMCID: PMC4674669
- DOI: 10.1038/ncomms10069
Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci
Abstract
Genome-wide association studies have been tremendously successful in identifying genetic variants associated with complex diseases. The majority of association signals are intergenic and evidence is accumulating that a high proportion of signals lie in enhancer regions. We use Capture Hi-C to investigate, for the first time, the interactions between associated variants for four autoimmune diseases and their functional targets in B- and T-cell lines. Here we report numerous looping interactions and provide evidence that only a minority of interactions are common to both B- and T-cell lines, suggesting interactions may be highly cell-type specific; some disease-associated SNPs do not interact with the nearest gene but with more compelling candidate genes (for example, FOXO1, AZI2) often situated several megabases away; and finally, regions associated with different autoimmune diseases interact with each other and the same promoter suggesting common autoimmune gene targets (for example, PTPRC, DEXI and ZFP36L1).
Figures

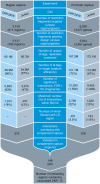
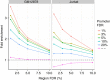


References
-
- Schoenfelder S., Clay I. & Fraser P. The transcriptional interactome: gene expression in 3D. Curr. Opin. Genet. Dev. 20, 127–133 (2010). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

