Plasma 24-metabolite Panel Predicts Preclinical Transition to Clinical Stages of Alzheimer's Disease
- PMID: 26617567
- PMCID: PMC4642213
- DOI: 10.3389/fneur.2015.00237
Plasma 24-metabolite Panel Predicts Preclinical Transition to Clinical Stages of Alzheimer's Disease
Abstract
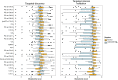
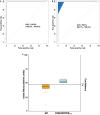
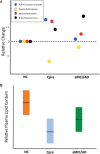
We recently documented plasma lipid dysregulation in preclinical late-onset Alzheimer's disease (LOAD). A 10 plasma lipid panel, predicted phenoconversion and provided 90% sensitivity and 85% specificity in differentiating an at-risk group from those that would remain cognitively intact. Despite these encouraging results, low positive predictive values limit the clinical usefulness of this panel as a screening tool in subjects aged 70-80 years or younger. In this report, we re-examine our metabolomic data, analyzing baseline plasma specimens from our group of phenoconverters (n = 28) and a matched set of cognitively normal subjects (n = 73), and discover and internally validate a panel of 24 plasma metabolites. The new panel provides a classifier with receiver operating characteristic area under the curve for the discovery and internal validation cohort of 1.0 and 0.995 (95% confidence intervals of 1.0-1.0, and 0.981-1.0), respectively. Twenty-two of the 24 metabolites were significantly dysregulated lipids. While positive and negative predictive values were improved compared to our 10-lipid panel, low positive predictive values provide a reality check on the utility of such biomarkers in this age group (or younger). Through inclusion of additional significantly dysregulated analyte species, our new biomarker panel provides greater accuracy in our cohort but remains limited by predictive power. Unfortunately, the novel metabolite panel alone may not provide improvement in counseling and management of at-risk individuals but may further improve selection of subjects for LOAD secondary prevention trials. We expect that external validation will remain challenging due to our stringent study design, especially compared with more diverse subject cohorts. We do anticipate, however, external validation of reduced plasma lipid species as a predictor of phenoconversion to either prodromal or manifest LOAD.
Keywords: Alzheimer’s disease; biomarkers; economics; ethics; lipids; metabolomics; risk assessment.
Figures

References
-
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement (2011) 7:280–92.10.1016/j.jalz.2011.03.003 - DOI - PMC - PubMed
-
- Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem (1993) 39:561–77. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

