Non-Coding RNA: Sequence-Specific Guide for Chromatin Modification and DNA Damage Signaling
- PMID: 26617633
- PMCID: PMC4643122
- DOI: 10.3389/fgene.2015.00320
Non-Coding RNA: Sequence-Specific Guide for Chromatin Modification and DNA Damage Signaling
Abstract
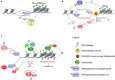
Chromatin conformation shapes the environment in which our genome is transcribed into RNA. Transcription is a source of DNA damage, thus it often occurs concomitantly to DNA damage signaling. Growing amounts of evidence suggest that different types of RNAs can, independently from their protein-coding properties, directly affect chromatin conformation, transcription and splicing, as well as promote the activation of the DNA damage response (DDR) and DNA repair. Therefore, transcription paradoxically functions to both threaten and safeguard genome integrity. On the other hand, DNA damage signaling is known to modulate chromatin to suppress transcription of the surrounding genetic unit. It is thus intriguing to understand how transcription can modulate DDR signaling while, in turn, DDR signaling represses transcription of chromatin around the DNA lesion. An unexpected player in this field is the RNA interference (RNAi) machinery, which play roles in transcription, splicing and chromatin modulation in several organisms. Non-coding RNAs (ncRNAs) and several protein factors involved in the RNAi pathway are well known master regulators of chromatin while only recent reports show their involvement in DDR. Here, we discuss the experimental evidence supporting the idea that ncRNAs act at the genomic loci from which they are transcribed to modulate chromatin, DDR signaling and DNA repair.
Keywords: DNA-damage response; RNA interference; chromatin modulation; non-coding RNA; transcription.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

