Mapping the Free Energy Landscape of PKA Inhibition and Activation: A Double-Conformational Selection Model for the Tandem cAMP-Binding Domains of PKA RIα
- PMID: 26618408
- PMCID: PMC4664472
- DOI: 10.1371/journal.pbio.1002305
Mapping the Free Energy Landscape of PKA Inhibition and Activation: A Double-Conformational Selection Model for the Tandem cAMP-Binding Domains of PKA RIα
Abstract
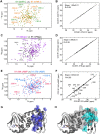
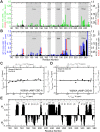
Protein Kinase A (PKA) is the major receptor for the cyclic adenosine monophosphate (cAMP) secondary messenger in eukaryotes. cAMP binds to two tandem cAMP-binding domains (CBD-A and -B) within the regulatory subunit of PKA (R), unleashing the activity of the catalytic subunit (C). While CBD-A in RIα is required for PKA inhibition and activation, CBD-B functions as a "gatekeeper" domain that modulates the control exerted by CBD-A. Preliminary evidence suggests that CBD-B dynamics are critical for its gatekeeper function. To test this hypothesis, here we investigate by Nuclear Magnetic Resonance (NMR) the two-domain construct RIα (91-379) in its apo, cAMP2, and C-bound forms. Our comparative NMR analyses lead to a double conformational selection model in which each apo CBD dynamically samples both active and inactive states independently of the adjacent CBD within a nearly degenerate free energy landscape. Such degeneracy is critical to explain the sensitivity of CBD-B to weak interactions with C and its high affinity for cAMP. Binding of cAMP eliminates this degeneracy, as it selectively stabilizes the active conformation within each CBD and inter-CBD contacts, which require both cAMP and W260. The latter is contributed by CBD-B and mediates capping of the cAMP bound to CBD-A. The inter-CBD interface is dispensable for intra-CBD conformational selection, but is indispensable for full activation of PKA as it occludes C-subunit recognition sites within CBD-A. In addition, the two structurally homologous cAMP-bound CBDs exhibit marked differences in their residual dynamics profiles, supporting the notion that conservation of structure does not necessarily imply conservation of dynamics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12: 258–266. - PubMed
-
- Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101: 2381–2411. - PubMed
-
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9: 576–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

