RBFox1-mediated RNA splicing regulates cardiac hypertrophy and heart failure
- PMID: 26619120
- PMCID: PMC4701548
- DOI: 10.1172/JCI84015
RBFox1-mediated RNA splicing regulates cardiac hypertrophy and heart failure
Abstract
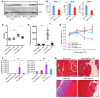
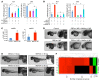
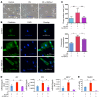
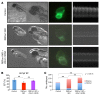
RNA splicing is a major contributor to total transcriptome complexity; however, the functional role and regulation of splicing in heart failure remain poorly understood. Here, we used a total transcriptome profiling and bioinformatic analysis approach and identified a muscle-specific isoform of an RNA splicing regulator, RBFox1 (also known as A2BP1), as a prominent regulator of alternative RNA splicing during heart failure. Evaluation of developing murine and zebrafish hearts revealed that RBFox1 is induced during postnatal cardiac maturation. However, we found that RBFox1 is markedly diminished in failing human and mouse hearts. In a mouse model, RBFox1 deficiency in the heart promoted pressure overload-induced heart failure. We determined that RBFox1 is a potent regulator of RNA splicing and is required for a conserved splicing process of transcription factor MEF2 family members that yields different MEF2 isoforms with differential effects on cardiac hypertrophic gene expression. Finally, induction of RBFox1 expression in murine pressure overload models substantially attenuated cardiac hypertrophy and pathological manifestations. Together, this study identifies regulation of RNA splicing by RBFox1 as an important player in transcriptome reprogramming during heart failure that influence pathogenesis of the disease.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL108186/HL/NHLBI NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- R01 HL122737/HL/NHLBI NIH HHS/United States
- R21 HL110667/HL/NHLBI NIH HHS/United States
- R01 HL103205/HL/NHLBI NIH HHS/United States
- HL103205/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- R01 HL115238/HL/NHLBI NIH HHS/United States
- HL110667/HL/NHLBI NIH HHS/United States
- U01HG007013/HG/NHGRI NIH HHS/United States
- R01HG006264/HG/NHGRI NIH HHS/United States
- R01 HG006264/HG/NHGRI NIH HHS/United States
- R01 HL070079/HL/NHLBI NIH HHS/United States
- HL070079/HL/NHLBI NIH HHS/United States
- HL115238/HL/NHLBI NIH HHS/United States
- U01 HG007013/HG/NHGRI NIH HHS/United States
- HL096980/HL/NHLBI NIH HHS/United States
- T32 HL069766/HL/NHLBI NIH HHS/United States
- R01 HL096980/HL/NHLBI NIH HHS/United States
- T32 HL-69766/HL/NHLBI NIH HHS/United States
- HL108186/HL/NHLBI NIH HHS/United States
- R01 GM049369/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

