Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by Entamoeba and Subsistence
- PMID: 26619199
- PMCID: PMC4664238
- DOI: 10.1371/journal.pgen.1005658
Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by Entamoeba and Subsistence
Abstract
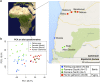
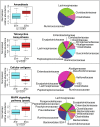
The human gut microbiota is impacted by host nutrition and health status and therefore represents a potentially adaptive phenotype influenced by metabolic and immune constraints. Previous studies contrasting rural populations in developing countries to urban industrialized ones have shown that industrialization is strongly correlated with patterns in human gut microbiota; however, we know little about the relative contribution of factors such as climate, diet, medicine, hygiene practices, host genetics, and parasitism. Here, we focus on fine-scale comparisons of African rural populations in order to (i) contrast the gut microbiota of populations inhabiting similar environments but having different traditional subsistence modes and either shared or distinct genetic ancestry, and (ii) examine the relationship between gut parasites and bacterial communities. Characterizing the fecal microbiota of Pygmy hunter-gatherers as well as Bantu individuals from both farming and fishing populations in Southwest Cameroon, we found that the gut parasite Entamoeba is significantly correlated with microbiome composition and diversity. We show that across populations, colonization by this protozoa can be predicted with 79% accuracy based on the composition of an individual's gut microbiota, and that several of the taxa most important for distinguishing Entamoeba absence or presence are signature taxa for autoimmune disorders. We also found gut communities to vary significantly with subsistence mode, notably with some taxa previously shown to be enriched in other hunter-gatherers groups (in Tanzania and Peru) also discriminating hunter-gatherers from neighboring farming or fishing populations in Cameroon.
Conflict of interest statement
The authors declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

