Transcriptional Regulation of Cystathionine-γ-Lyase in Endothelial Cells by NADPH Oxidase 4-Dependent Signaling
- PMID: 26620565
- PMCID: PMC4722457
- DOI: 10.1074/jbc.M115.685578
Transcriptional Regulation of Cystathionine-γ-Lyase in Endothelial Cells by NADPH Oxidase 4-Dependent Signaling
Abstract
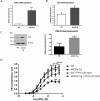
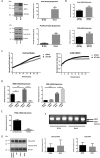
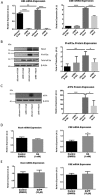
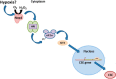
The gasotransmitter, hydrogen sulfide (H2S) is recognized as an important mediator of endothelial cell homeostasis and function that impacts upon vascular tone and blood pressure. Cystathionine-γ-lyase (CSE) is the predominant endothelial generator of H2S, and recent evidence suggests that its transcriptional expression is regulated by the reactive oxygen species, H2O2. However, the cellular source of H2O2 and the redox-dependent molecular signaling pathway that modulates this is not known. We aimed to investigate the role of Nox4, an endothelial generator of H2O2, in the regulation of CSE in endothelial cells. Both gain- and loss-of-function experiments in human endothelial cells in vitro demonstrated Nox4 to be a positive regulator of CSE transcription and protein expression. We demonstrate that this is dependent upon a heme-regulated inhibitor kinase/eIF2α/activating transcription factor 4 (ATF4) signaling module. ATF4 was further demonstrated to bind directly to cis-regulatory sequences within the first intron of CSE to activate transcription. Furthermore, CSE expression was also increased in cardiac microvascular endothelial cells, isolated from endothelial-specific Nox4 transgenic mice, compared with wild-type littermate controls. Using wire myography we demonstrate that endothelial-specific Nox4 transgenic mice exhibit a hypo-contractile phenotype in response to phenylephrine that was abolished when vessels were incubated with a CSE inhibitor, propargylglycine. We, therefore, conclude that Nox4 is a positive transcriptional regulator of CSE in endothelial cells and propose that it may in turn contribute to the regulation of vascular tone via the modulation of H2S production.
Keywords: ATF4; CSE; NADPH oxidase 4; endothelial cell; hydrogen sulfide; reactive oxygen species (ROS); redox signaling; transcription regulation; transgenic mice; vascular tone.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Dysregulation of cystathionine γ-lyase (CSE)/hydrogen sulfide pathway contributes to ox-LDL-induced inflammation in macrophage.Cell Signal. 2013 Nov;25(11):2255-62. doi: 10.1016/j.cellsig.2013.07.010. Epub 2013 Jul 18. Cell Signal. 2013. PMID: 23872072
-
Endogenous Hydrogen Sulfide Ameliorates NOX4 Induced Oxidative Stress in LPS-Stimulated Macrophages and Mice.Cell Physiol Biochem. 2018;47(2):458-474. doi: 10.1159/000489980. Epub 2018 May 22. Cell Physiol Biochem. 2018. PMID: 29794432
-
Angiotensin II downregulates vascular endothelial cell hydrogen sulfide production by enhancing cystathionine γ-lyase degradation through ROS-activated ubiquitination pathway.Biochem Biophys Res Commun. 2019 Jun 30;514(3):907-912. doi: 10.1016/j.bbrc.2019.05.021. Epub 2019 May 10. Biochem Biophys Res Commun. 2019. PMID: 31084929
-
Regulation of cystathionine gamma-lyase/H₂S system and its pathological implication.Front Biosci (Landmark Ed). 2014 Jun 1;19(8):1355-69. doi: 10.2741/4286. Front Biosci (Landmark Ed). 2014. PMID: 24896355 Review.
-
Hydrogen sulfide generation in mammals: the molecular biology of cystathionine-β- synthase (CBS) and cystathionine-γ-lyase (CSE).Inflamm Allergy Drug Targets. 2011 Apr;10(2):85-91. doi: 10.2174/187152811794776286. Inflamm Allergy Drug Targets. 2011. PMID: 21275900 Review.
Cited by
-
Why Is Iron Deficiency/Anemia Linked to Alzheimer's Disease and Its Comorbidities, and How Is It Prevented?Biomedicines. 2023 Aug 30;11(9):2421. doi: 10.3390/biomedicines11092421. Biomedicines. 2023. PMID: 37760862 Free PMC article. Review.
-
Cystathionine γ-lyase protects vascular endothelium: a role for inhibition of histone deacetylase 6.Am J Physiol Heart Circ Physiol. 2017 Apr 1;312(4):H711-H720. doi: 10.1152/ajpheart.00724.2016. Epub 2017 Feb 10. Am J Physiol Heart Circ Physiol. 2017. PMID: 28188215 Free PMC article.
-
Vascular biology of hydrogen sulfide.Am J Physiol Cell Physiol. 2017 May 1;312(5):C537-C549. doi: 10.1152/ajpcell.00329.2016. Epub 2017 Feb 1. Am J Physiol Cell Physiol. 2017. PMID: 28148499 Free PMC article. Review.
-
An Update on Hydrogen Sulfide and Nitric Oxide Interactions in the Cardiovascular System.Oxid Med Cell Longev. 2018 Sep 9;2018:4579140. doi: 10.1155/2018/4579140. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 30271527 Free PMC article. Review.
-
Decreased vascular H2S production is associated with vascular oxidative stress in rats fed a high-fat western diet.Naunyn Schmiedebergs Arch Pharmacol. 2016 Jul;389(7):783-90. doi: 10.1007/s00210-016-1244-4. Epub 2016 Apr 18. Naunyn Schmiedebergs Arch Pharmacol. 2016. PMID: 27087304
References
-
- Aird W. C. (2007) Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ. Res. 100, 158–173 - PubMed
-
- Mustafa A. K., Sikka G., Gazi S. K., Steppan J., Jung S. M., Bhunia A. K., Barodka V. M., Gazi F. K., Barrow R. K., Wang R., Amzel L. M., Berkowitz D. E., and Snyder S. H. (2011) Hydrogen sulfide as endothelium-derived hyperpolarizing factor sulfhydrates potassium channels. Circ. Res. 109, 1259–1268 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RG/12/12/29872/BHF_/British Heart Foundation/United Kingdom
- G0700320/MRC_/Medical Research Council/United Kingdom
- MR/K003232/1/MRC_/Medical Research Council/United Kingdom
- BB/C503646/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0600785/MRC_/Medical Research Council/United Kingdom
- G1000458/MRC_/Medical Research Council/United Kingdom
- FS/11/45/28859/BHF_/British Heart Foundation/United Kingdom
- FS/11/80/29330/BHF_/British Heart Foundation/United Kingdom
- PG/13/13/30018/BHF_/British Heart Foundation/United Kingdom
- RG/13/11/30384/BHF_/British Heart Foundation/United Kingdom
- MR/L009684/1/MRC_/Medical Research Council/United Kingdom
- PG/10/98/28655/BHF_/British Heart Foundation/United Kingdom
- RE/13/2/30182/BHF_/British Heart Foundation/United Kingdom
- PG/15/26/31373/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources

