Branched-chain amino acid catabolism is a conserved regulator of physiological ageing
- PMID: 26620638
- PMCID: PMC4686672
- DOI: 10.1038/ncomms10043
Branched-chain amino acid catabolism is a conserved regulator of physiological ageing
Abstract
Ageing has been defined as a global decline in physiological function depending on both environmental and genetic factors. Here we identify gene transcripts that are similarly regulated during physiological ageing in nematodes, zebrafish and mice. We observe the strongest extension of lifespan when impairing expression of the branched-chain amino acid transferase-1 (bcat-1) gene in C. elegans, which leads to excessive levels of branched-chain amino acids (BCAAs). We further show that BCAAs reduce a LET-363/mTOR-dependent neuro-endocrine signal, which we identify as DAF-7/TGFβ, and that impacts lifespan depending on its related receptors, DAF-1 and DAF-4, as well as ultimately on DAF-16/FoxO and HSF-1 in a cell-non-autonomous manner. The transcription factor HLH-15 controls and epistatically synergizes with BCAT-1 to modulate physiological ageing. Lastly and consistent with previous findings in rodents, nutritional supplementation of BCAAs extends nematodal lifespan. Taken together, BCAAs act as periphery-derived metabokines that induce a central neuro-endocrine response, culminating in extended healthspan.
Figures

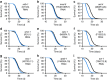
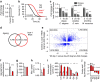
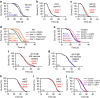
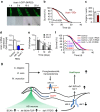
References
-
- McCay C. M., Crowel M. F. & Maynard L. A. The effect of retarded growth upon the length of the life span and upon ultimate body size. J. Nutr. 10, 63–79 (1935). - PubMed
-
- Klass M. R. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech. Ageing Dev. 22, 279–286 (1983). - PubMed
-
- Kirkwood T. B. & Melov S. On the programmed/non-programmed nature of ageing within the life history. Curr. Biol. 21, R701–R707 (2011). - PubMed
-
- Gems D. What is an anti-aging treatment? Exp. Gerontol. 58C, 14–18 (2014). - PubMed
-
- Johnson T. E. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science 249, 908–912 (1990). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

