Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal
- PMID: 26620920
- PMCID: PMC4697335
- DOI: 10.1038/ncomms10062
Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal
Abstract
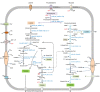
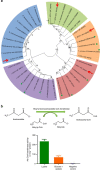
Human intestinal bacteria produce butyrate, which has signalling properties and can be used as energy source by enterocytes thus influencing colonic health. However, the pathways and the identity of bacteria involved in this process remain unclear. Here we describe the isolation from the human intestine of Intestinimonas strain AF211, a bacterium that can convert lysine stoichiometrically into butyrate and acetate when grown in a synthetic medium. Intestinimonas AF211 also converts the Amadori product fructoselysine, which is abundantly formed in heated foods via the Maillard reaction, into butyrate. The butyrogenic pathway includes a specific CoA transferase that is overproduced during growth on lysine. Bacteria related to Intestinimonas AF211 as well as the genetic coding capacity for fructoselysine conversion are abundantly present in colonic samples from some healthy human subjects. Our results indicate that protein can serve as a source of butyrate in the human colon, and its conversion by Intestinimonas AF211 and related butyrogens may protect the host from the undesired side effects of Amadori reaction products.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

