Autism-associated R451C mutation in neuroligin3 leads to activation of the unfolded protein response in a PC12 Tet-On inducible system
- PMID: 26621873
- PMCID: PMC4747159
- DOI: 10.1042/BJ20150274
Autism-associated R451C mutation in neuroligin3 leads to activation of the unfolded protein response in a PC12 Tet-On inducible system
Abstract
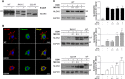
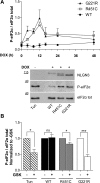
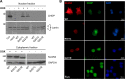
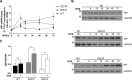
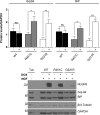
Several forms of monogenic heritable autism spectrum disorders are associated with mutations in the neuroligin genes. The autism-linked substitution R451C in neuroligin3 induces local misfolding of its extracellular domain, causing partial retention in the ER (endoplasmic reticulum) of expressing cells. We have generated a PC12 Tet-On cell model system with inducible expression of wild-type or R451C neuroligin3 to investigate whether there is activation of the UPR (unfolded protein response) as a result of misfolded protein retention. As a positive control for protein misfolding, we also expressed the mutant G221R neuroligin3, which is known to be completely retained within the ER. Our data show that overexpression of either R451C or G221R mutant proteins leads to the activation of all three signalling branches of the UPR downstream of the stress sensors ATF6 (activating transcription factor 6), IRE1 (inositol-requiring enzyme 1) and PERK [PKR (dsRNA-dependent protein kinase)-like endoplasmic reticulum kinase]. Each branch displayed different activation profiles that partially correlated with the degree of misfolding caused by each mutation. We also show that up-regulation of BiP (immunoglobulin heavy-chain-binding protein) and CHOP [C/EBP (CCAAT/enhancer-binding protein)-homologous protein] was induced by both mutant proteins but not by wild-type neuroligin3, both in proliferative cells and cells differentiated to a neuron-like phenotype. Collectively, our data show that mutant R451C neuroligin3 activates the UPR in a novel cell model system, suggesting that this cellular response may have a role in monogenic forms of autism characterized by misfolding mutations.
Keywords: ER stress; autism; molecular chaperones; neuroligin; protein misfolding; unfolded protein response.
© 2016 Authors.
Figures


Similar articles
-
UPR activation specifically modulates glutamate neurotransmission in the cerebellum of a mouse model of autism.Neurobiol Dis. 2018 Dec;120:139-150. doi: 10.1016/j.nbd.2018.08.026. Epub 2018 Sep 7. Neurobiol Dis. 2018. PMID: 30201312
-
Neuroligin trafficking deficiencies arising from mutations in the alpha/beta-hydrolase fold protein family.J Biol Chem. 2010 Sep 10;285(37):28674-82. doi: 10.1074/jbc.M110.139519. Epub 2010 Jul 8. J Biol Chem. 2010. PMID: 20615874 Free PMC article.
-
Folding anomalies of neuroligin3 caused by a mutation in the alpha/beta-hydrolase fold domain.Chem Biol Interact. 2010 Sep 6;187(1-3):56-8. doi: 10.1016/j.cbi.2010.03.012. Epub 2010 Mar 12. Chem Biol Interact. 2010. PMID: 20227402 Free PMC article.
-
Endoplasmic reticulum stress and eIF2α phosphorylation: The Achilles heel of pancreatic β cells.Mol Metab. 2017 Jul 12;6(9):1024-1039. doi: 10.1016/j.molmet.2017.06.001. eCollection 2017 Sep. Mol Metab. 2017. PMID: 28951826 Free PMC article. Review.
-
Unraveling the Role of Neuroligin3 in Autism Spectrum Disorders: Pathophysiological Insights and Targeted Therapies.CNS Neurol Disord Drug Targets. 2024;23(7):801-811. doi: 10.2174/1871527323666230727102244. CNS Neurol Disord Drug Targets. 2024. PMID: 37497709 Review.
Cited by
-
Sleep deficiency as a driver of cellular stress and damage in neurological disorders.Sleep Med Rev. 2022 Jun;63:101616. doi: 10.1016/j.smrv.2022.101616. Epub 2022 Feb 26. Sleep Med Rev. 2022. PMID: 35381445 Free PMC article. Review.
-
The Unfolded Protein Response and Autophagy as Drug Targets in Neuropsychiatric Disorders.Front Cell Neurosci. 2020 Sep 29;14:554548. doi: 10.3389/fncel.2020.554548. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33132844 Free PMC article. Review.
-
Understanding the Unfolded Protein Response (UPR) Pathway: Insights into Neuropsychiatric Disorders and Therapeutic Potentials.Biomol Ther (Seoul). 2024 Mar 1;32(2):183-191. doi: 10.4062/biomolther.2023.181. Biomol Ther (Seoul). 2024. PMID: 38410073 Free PMC article. Review.
-
A Perspective on the Potential Involvement of Impaired Proteostasis in Neuropsychiatric Disorders.Biol Psychiatry. 2022 Feb 15;91(4):335-345. doi: 10.1016/j.biopsych.2021.09.001. Epub 2021 Sep 14. Biol Psychiatry. 2022. PMID: 34836635 Free PMC article. Review.
-
Cellular stress and apoptosis contribute to the pathogenesis of autism spectrum disorder.Autism Res. 2018 Jul;11(7):1076-1090. doi: 10.1002/aur.1966. Epub 2018 May 15. Autism Res. 2018. PMID: 29761862 Free PMC article.
References
-
- Jamain S., Quach H., Betancur C., Rastam M., Colineaux C., Gillberg I.C., Soderstrom H., Giros B., Leboyer M., Gillberg C., Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

