Automated physics-based design of synthetic riboswitches from diverse RNA aptamers
- PMID: 26621913
- PMCID: PMC4705656
- DOI: 10.1093/nar/gkv1289
Automated physics-based design of synthetic riboswitches from diverse RNA aptamers
Abstract
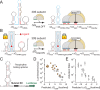
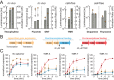
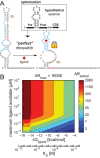
Riboswitches are shape-changing regulatory RNAs that bind chemicals and regulate gene expression, directly coupling sensing to cellular actuation. However, it remains unclear how their sequence controls the physics of riboswitch switching and activation, particularly when changing the ligand-binding aptamer domain. We report the development of a statistical thermodynamic model that predicts the sequence-structure-function relationship for translation-regulating riboswitches that activate gene expression, characterized inside cells and within cell-free transcription-translation assays. Using the model, we carried out automated computational design of 62 synthetic riboswitches that used six different RNA aptamers to sense diverse chemicals (theophylline, tetramethylrosamine, fluoride, dopamine, thyroxine, 2,4-dinitrotoluene) and activated gene expression by up to 383-fold. The model explains how aptamer structure, ligand affinity, switching free energy and macromolecular crowding collectively control riboswitch activation. Our model-based approach for engineering riboswitches quantitatively confirms several physical mechanisms governing ligand-induced RNA shape-change and enables the development of cell-free and bacterial sensors for diverse applications.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Mandal M., Breaker R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. - PubMed
-
- Montange R.K., Batey R.T. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. - PubMed
-
- Berens C., Suess B. Riboswitch engineering—making the all-important second and third steps. Curr. Opin. Biotechnol. 2015;31:10–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

