Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014
- PMID: 26625712
- PMCID: PMC4686777
- DOI: 10.1038/ncomms10061
Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014
Abstract
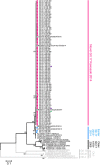
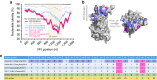
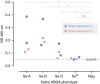
Norovirus genogroup II genotype 4 (GII.4) has been the predominant cause of viral gastroenteritis since 1996. Here we show that during the winter of 2014-2015, an emergent variant of a previously rare norovirus GII.17 genotype, Kawasaki 2014, predominated in Hong Kong and outcompeted contemporary GII.4 Sydney 2012 in hospitalized cases. GII.17 cases were significantly older than GII.4 cases. Root-to-tip and Bayesian BEAST analyses estimate GII.17 viral protein 1 (VP1) evolves one order of magnitude faster than GII.4 VP1. Residue substitutions and insertion occur in four of five inferred antigenic epitopes, suggesting immune evasion. Sequential GII.4-GII.17 infections are noted, implicating a lack of cross-protection. Virus bound to saliva of secretor histo-blood groups A, B and O, indicating broad susceptibility. This fast-evolving, broadly recognizing and probably immune-escaped emergent GII.17 variant causes severe gastroenteritis and hospitalization across all age groups, including populations who were previously less vulnerable to GII.4 variants; therefore, the global spread of GII.17 Kawasaki 2014 needs to be monitored.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

