High level of inbreeding in final phase of 1000 Genomes Project
- PMID: 26625947
- PMCID: PMC4667178
- DOI: 10.1038/srep17453
High level of inbreeding in final phase of 1000 Genomes Project
Abstract
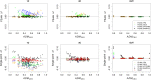
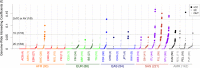
The 1000 Genomes Project provides a unique source of whole genome sequencing data for studies of human population genetics and human diseases. The last release of this project includes more than 2,500 sequenced individuals from 26 populations. Although relationships among individuals have been investigated in some of the populations, inbreeding has never been studied. In this article, we estimated the genomic inbreeding coefficient of each individual and found an unexpected high level of inbreeding in 1000 Genomes data: nearly a quarter of the individuals were inbred and around 4% of them had inbreeding coefficients similar or greater than the ones expected for first-cousin offspring. Inbred individuals were found in each of the 26 populations, with some populations showing proportions of inbred individuals above 50%. We also detected 227 previously unreported pairs of close relatives (up to and including first-cousins). Thus, we propose subsets of unrelated and outbred individuals, for use by the scientific community. In addition, because admixed populations are present in the 1000 Genomes Project, we performed simulations to study the robustness of inbreeding coefficient estimates in the presence of admixture. We found that our multi-point approach (FSuite) was quite robust to admixture, unlike single-point methods (PLINK).
Figures
References
-
- Grossman S. R. et al.. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327, 883–886 (2010). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

