Accuracy of genomic selection for alfalfa biomass yield in different reference populations
- PMID: 26626170
- PMCID: PMC4667460
- DOI: 10.1186/s12864-015-2212-y
Accuracy of genomic selection for alfalfa biomass yield in different reference populations
Abstract
Background: Genomic selection based on genotyping-by-sequencing (GBS) data could accelerate alfalfa yield gains, if it displayed moderate ability to predict parent breeding values. Its interest would be enhanced by predicting ability also for germplasm/reference populations other than those for which it was defined. Predicting accuracy may be influenced by statistical models, SNP calling procedures and missing data imputation strategies.
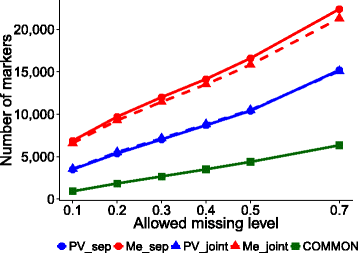
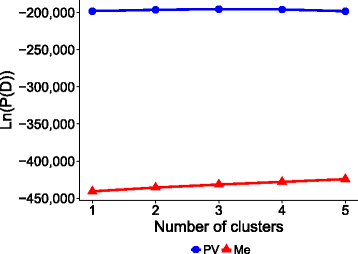
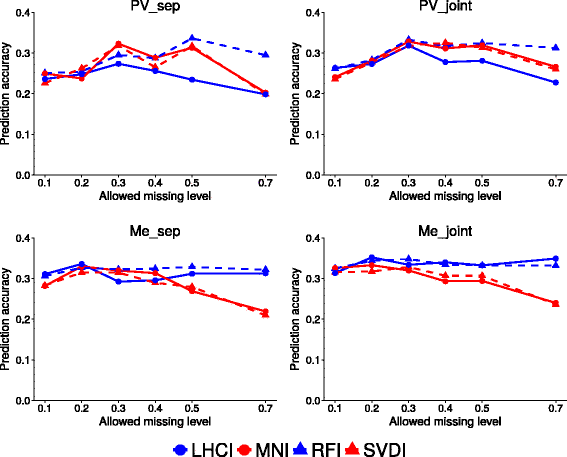
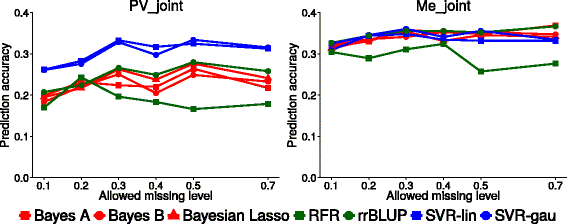
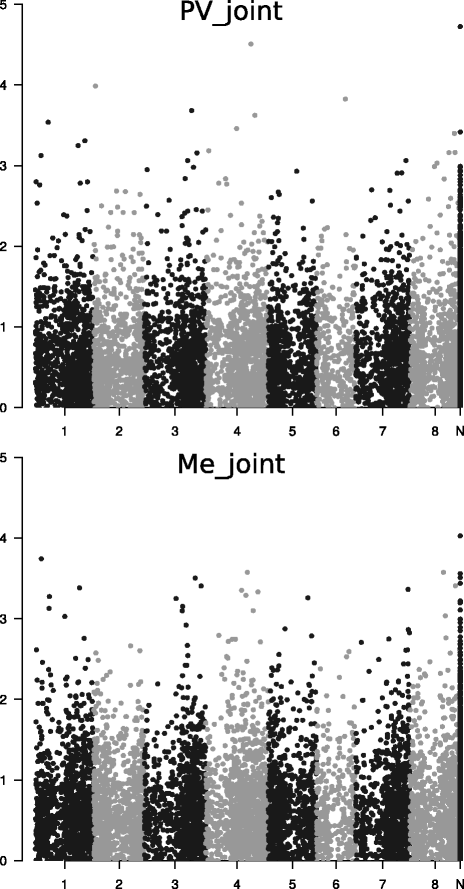
Results: Landrace and variety material from two genetically-contrasting reference populations, i.e., 124 elite genotypes adapted to the Po Valley (sub-continental climate; PV population) and 154 genotypes adapted to Mediterranean-climate environments (Me population), were genotyped by GBS and phenotyped in separate environments for dry matter yield of their dense-planted half-sib progenies. Both populations showed no sub-population genetic structure. Predictive accuracy was higher by joint rather than separate SNP calling for the two data sets, and using random forest imputation of missing data. Highest accuracy was obtained using Support Vector Regression (SVR) for PV, and Ridge Regression BLUP and SVR for Me germplasm. Bayesian methods (Bayes A, Bayes B and Bayesian Lasso) tended to be less accurate. Random Forest Regression was the least accurate model. Accuracy attained about 0.35 for Me in the range of 0.30-0.50 missing data, and 0.32 for PV at 0.50 missing data, using at least 10,000 SNP markers. Cross-population predictions based on a smaller subset of common SNPs implied a relative loss of accuracy of about 25% for Me and 30% for PV. Genome-wide association analyses based on large subsets of M. truncatula-aligned markers revealed many SNPs with modest association with yield, and some genome areas hosting putative QTLs. A comparison of genomic vs. conventional selection for parent breeding value assuming 1-year vs. 5-year selection cycles, respectively, indicated over three-fold greater predicted yield gain per unit time for genomic selection.
Conclusions: Genomic selection for alfalfa yield is promising, based on its moderate prediction accuracy, moderate value of cross-population predictions, and lack of sub-population structure. There is limited scope for searching individual QTLs with overwhelming effect on yield. Some of our results can contribute to better design of genomic selection experiments for alfalfa and other crops with similar mating systems.
Figures

References
-
- Annicchiarico P. Alfalfa forage yield and leaf/stem ratio: narrow-sense heritability, genetic correlation, and parent selection procedures. Euphytica. 2015;205:409–20. doi: 10.1007/s10681-015-1399-y. - DOI
-
- Lamb JF, Jung H-JG, Riday H. Growth environment, harvest management and germplasm impacts on potential ethanol and crude protein yield in alfalfa. Biomass Bioenergy. 2014;63:114–25. doi: 10.1016/j.biombioe.2014.02.006. - DOI
-
- Annicchiarico P, Scotti C, Carelli M, Pecetti L. Questions and avenues for lucerne improvement. Czech J Genet Plant Breed. 2010;46:1–13.
-
- Annicchiarico P, Barrett B, Brummer EC, Julier B, Marshall AH. Achievements and challenges in improving temperate perennial forage legumes. Crit Rev Plant Sci. 2015;34:327–80. doi: 10.1080/07352689.2014.898462. - DOI
-
- Li X, Brummer EC. Applied genetics and genomics in alfalfa breeding. Agron. 2012;2:40–61. doi: 10.3390/agronomy2010040. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

