PC-TraFF: identification of potentially collaborating transcription factors using pointwise mutual information
- PMID: 26627005
- PMCID: PMC4667426
- DOI: 10.1186/s12859-015-0827-2
PC-TraFF: identification of potentially collaborating transcription factors using pointwise mutual information
Abstract
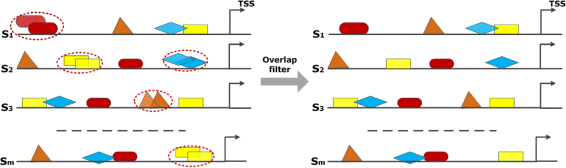
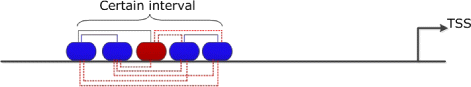
Background: Transcription factors (TFs) are important regulatory proteins that govern transcriptional regulation. Today, it is known that in higher organisms different TFs have to cooperate rather than acting individually in order to control complex genetic programs. The identification of these interactions is an important challenge for understanding the molecular mechanisms of regulating biological processes. In this study, we present a new method based on pointwise mutual information, PC-TraFF, which considers the genome as a document, the sequences as sentences, and TF binding sites (TFBSs) as words to identify interacting TFs in a set of sequences.
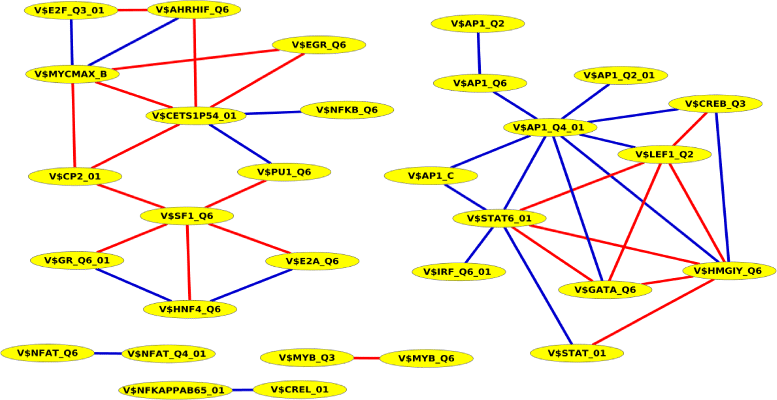
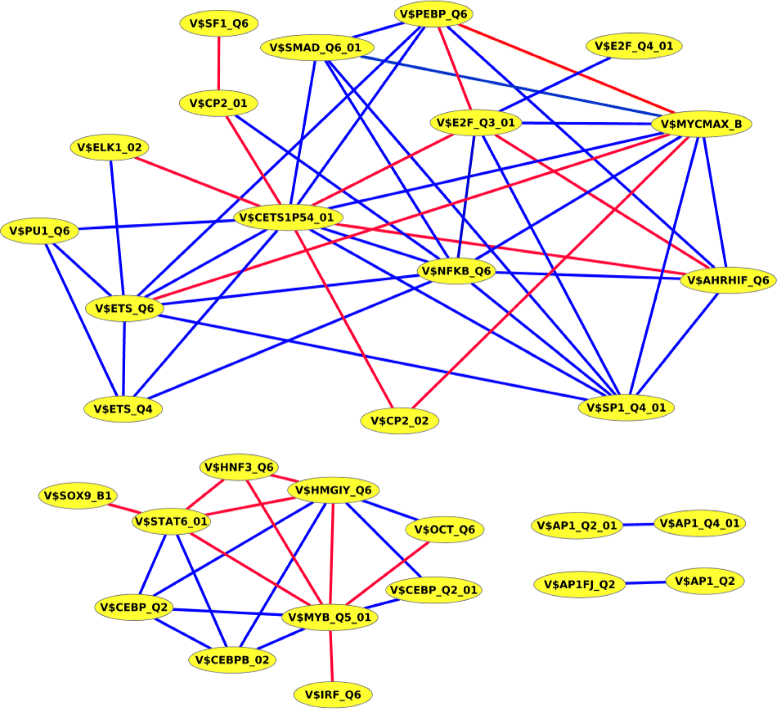
Results: To demonstrate the effectiveness of PC-TraFF, we performed a genome-wide analysis and a breast cancer-associated sequence set analysis for protein coding and miRNA genes. Our results show that in any of these sequence sets, PC-TraFF is able to identify important interacting TF pairs, for most of which we found support by previously published experimental results. Further, we made a pairwise comparison between PC-TraFF and three conventional methods. The outcome of this comparison study strongly suggests that all these methods focus on different important aspects of interaction between TFs and thus the pairwise overlap between any of them is only marginal.
Conclusions: In this study, adopting the idea from the field of linguistics in the field of bioinformatics, we develop a new information theoretic method, PC-TraFF, for the identification of potentially collaborating transcription factors based on the idiosyncrasy of their binding site distributions on the genome. The results of our study show that PC-TraFF can succesfully identify known interacting TF pairs and thus its currently biologically uncorfirmed predictions could provide new hypotheses for further experimental validation. Additionally, the comparison of the results of PC-TraFF with the results of previous methods demonstrates that different methods with their specific scopes can perfectly supplement each other. Overall, our analyses indicate that PC-TraFF is a time-efficient method where its algorithm has a tractable computational time and memory consumption. The PC-TraFF server is freely accessible at http://pctraff.bioinf.med.uni-goettingen.de/.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

