Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations
- PMID: 26627007
- PMCID: PMC4681596
- DOI: 10.1158/0008-5472.CAN-15-1654
Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations
Abstract
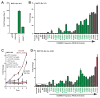
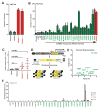
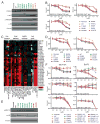
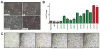
Large-scale sequencing efforts are uncovering the complexity of cancer genomes, which are composed of causal "driver" mutations that promote tumor progression along with many more pathologically neutral "passenger" events. The majority of mutations, both in known cancer drivers and uncharacterized genes, are generally of low occurrence, highlighting the need to functionally annotate the long tail of infrequent mutations present in heterogeneous cancers. Here we describe a mutation assessment pipeline enabled by high-throughput engineering of molecularly barcoded gene variant expression clones identified by tumor sequencing. We first used this platform to functionally assess tail mutations observed in PIK3CA, which encodes the catalytic subunit alpha of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) frequently mutated in cancer. Orthogonal screening for PIK3CA variant activity using in vitro and in vivo cell growth and transformation assays differentiated driver from passenger mutations, revealing that PIK3CA variant activity correlates imperfectly with its mutation frequency across breast cancer populations. Although PIK3CA mutations with frequencies above 5% were significantly more oncogenic than wild-type in all assays, mutations occurring at 0.07% to 5.0% included those with and without oncogenic activities that ranged from weak to strong in at least one assay. Proteomic profiling coupled with therapeutic sensitivity assays on PIK3CA variant-expressing cell models revealed variant-specific activation of PI3K signaling as well as other pathways that include the MEK1/2 module of mitogen-activated protein kinase pathway. Our data indicate that cancer treatments will need to increasingly consider the functional relevance of specific mutations in driver genes rather than considering all mutations in drivers as equivalent.
©2015 American Association for Cancer Research.
Conflict of interest statement
Figures

References
-
- Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7(6):561–73. - PubMed
-
- Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65(23):10992–1000. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

