Brg1 coordinates multiple processes during retinogenesis and is a tumor suppressor in retinoblastoma
- PMID: 26628093
- PMCID: PMC4712833
- DOI: 10.1242/dev.124800
Brg1 coordinates multiple processes during retinogenesis and is a tumor suppressor in retinoblastoma
Abstract
Retinal development requires precise temporal and spatial coordination of cell cycle exit, cell fate specification, cell migration and differentiation. When this process is disrupted, retinoblastoma, a developmental tumor of the retina, can form. Epigenetic modulators are central to precisely coordinating developmental events, and many epigenetic processes have been implicated in cancer. Studying epigenetic mechanisms in development is challenging because they often regulate multiple cellular processes; therefore, elucidating the primary molecular mechanisms involved can be difficult. Here we explore the role of Brg1 (Smarca4) in retinal development and retinoblastoma in mice using molecular and cellular approaches. Brg1 was found to regulate retinal size by controlling cell cycle length, cell cycle exit and cell survival during development. Brg1 was not required for cell fate specification but was required for photoreceptor differentiation and cell adhesion/polarity programs that contribute to proper retinal lamination during development. The combination of defective cell differentiation and lamination led to retinal degeneration in Brg1-deficient retinae. Despite the hypocellularity, premature cell cycle exit, increased cell death and extended cell cycle length, retinal progenitor cells persisted in Brg1-deficient retinae, making them more susceptible to retinoblastoma. ChIP-Seq analysis suggests that Brg1 might regulate gene expression through multiple mechanisms.
Keywords: Epigenetics; Mouse; Retina development; Retinoblastoma; SWI/SNF.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
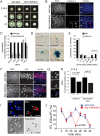
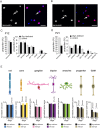
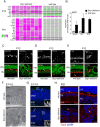



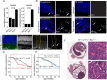
Similar articles
-
SWI/SNF chromatin-remodeling enzymes Brahma-related gene 1 (BRG1) and Brahma (BRM) are dispensable in multiple models of postnatal angiogenesis but are required for vascular integrity in infant mice.J Am Heart Assoc. 2015 Apr 22;4(4):e001972. doi: 10.1161/JAHA.115.001972. J Am Heart Assoc. 2015. PMID: 25904594 Free PMC article.
-
The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD.Development. 2005 Jan;132(1):105-15. doi: 10.1242/dev.01548. Epub 2004 Dec 2. Development. 2005. PMID: 15576411
-
The reversible epigenetic silencing of BRM: implications for clinical targeted therapy.Oncogene. 2007 Oct 25;26(49):7058-66. doi: 10.1038/sj.onc.1210514. Epub 2007 Jun 4. Oncogene. 2007. PMID: 17546055
-
The SWI/SNF complex and cancer.Oncogene. 2009 Apr 9;28(14):1653-68. doi: 10.1038/onc.2009.4. Epub 2009 Feb 23. Oncogene. 2009. PMID: 19234488 Review.
-
The RB protein family in retinal development and retinoblastoma: new insights from new mouse models.Dev Neurosci. 2004;26(5-6):417-34. doi: 10.1159/000082284. Dev Neurosci. 2004. PMID: 15855771 Review.
Cited by
-
Epigenetic control of gene regulation during development and disease: A view from the retina.Prog Retin Eye Res. 2018 Jul;65:1-27. doi: 10.1016/j.preteyeres.2018.03.002. Epub 2018 Mar 12. Prog Retin Eye Res. 2018. PMID: 29544768 Free PMC article. Review.
-
Building a Mammalian Retina: An Eye on Chromatin Structure.Front Genet. 2021 Oct 25;12:775205. doi: 10.3389/fgene.2021.775205. eCollection 2021. Front Genet. 2021. PMID: 34764989 Free PMC article. Review.
-
Deciphering the Retinal Epigenome during Development, Disease and Reprogramming: Advancements, Challenges and Perspectives.Cells. 2022 Feb 25;11(5):806. doi: 10.3390/cells11050806. Cells. 2022. PMID: 35269428 Free PMC article. Review.
-
Loss of BRG1 induces CRC cell senescence by regulating p53/p21 pathway.Cell Death Dis. 2017 Feb 9;8(2):e2607. doi: 10.1038/cddis.2017.1. Cell Death Dis. 2017. PMID: 28182012 Free PMC article.
-
Samd7 is a cell type-specific PRC1 component essential for establishing retinal rod photoreceptor identity.Proc Natl Acad Sci U S A. 2017 Sep 26;114(39):E8264-E8273. doi: 10.1073/pnas.1707021114. Epub 2017 Sep 12. Proc Natl Acad Sci U S A. 2017. PMID: 28900001 Free PMC article.
References
-
- Ajioka I., Martins R. A. P., Bayazitov I. T., Donovan S., Johnson D. A., Frase S., Cicero S. A., Boyd K., Zakharenko S. S. and Dyer M. A. (2007). Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 131, 378-390. 10.1016/j.cell.2007.09.036 - DOI - PMC - PubMed
-
- Attanasio C., Nord A. S., Zhu Y., Blow M. J., Biddie S. C., Mendenhall E. M., Dixon J., Wright C., Hosseini R., Akiyama J. A. et al. (2014). Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 24, 920-929. 10.1101/gr.168930.113 - DOI - PMC - PubMed
-
- Benavente C. A. , Finkelstein D. , Johnson D. A. , Marine J. C. , Ashery-Padan R. and Dyer M. A. (2014). Chromatin remodelers HELLS and UHRF1 mediate the epigenetic deregulation of genes that drive retinoblastoma tumor progression. Oncotarget 5, 9594-9608. 10.18632/oncotarget.2468 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 HG003988/HG/NHGRI NIH HHS/United States
- R01 EY018599/EY/NEI NIH HHS/United States
- U01 DE020060/DE/NIDCR NIH HHS/United States
- CA168875/CA/NCI NIH HHS/United States
- CA21765/CA/NCI NIH HHS/United States
- R01 EY014867/EY/NEI NIH HHS/United States
- R01 CA168875/CA/NCI NIH HHS/United States
- U54HG006997/HG/NHGRI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- Howard Hughes Medical Institute/United States
- EY018599/EY/NEI NIH HHS/United States
- EY014867/EY/NEI NIH HHS/United States
- R01HG003988/HG/NHGRI NIH HHS/United States
- U54 HG006997/HG/NHGRI NIH HHS/United States
- U01DE020060NIH/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

