The Binding Interface between Human APOBEC3F and HIV-1 Vif Elucidated by Genetic and Computational Approaches
- PMID: 26628363
- PMCID: PMC4684092
- DOI: 10.1016/j.celrep.2015.10.067
The Binding Interface between Human APOBEC3F and HIV-1 Vif Elucidated by Genetic and Computational Approaches
Abstract
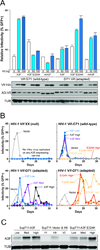
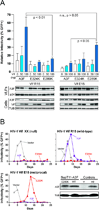
APOBEC3 family DNA cytosine deaminases provide overlapping defenses against pathogen infections. However, most viruses have elaborate evasion mechanisms such as the HIV-1 Vif protein, which subverts cellular CBF-β and a polyubiquitin ligase complex to neutralize these enzymes. Despite advances in APOBEC3 and Vif biology, a full understanding of this direct host-pathogen conflict has been elusive. We combine virus adaptation and computational studies to interrogate the APOBEC3F-Vif interface and build a robust structural model. A recurring compensatory amino acid substitution from adaptation experiments provided an initial docking constraint, and microsecond molecular dynamic simulations optimized interface contacts. Virus infectivity experiments validated a long-lasting electrostatic interaction between APOBEC3F E289 and HIV-1 Vif R15. Taken together with mutagenesis results, we propose a wobble model to explain how HIV-1 Vif has evolved to bind different APOBEC3 enzymes and, more generally, how pathogens may evolve to escape innate host defenses.
Keywords: APOBEC3F; APOBEC3F-Vif interface; HIV-1; Vif; pathogen-host interaction.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI083196/AI/NIAID NIH HHS/United States
- R37 AI064046/AI/NIAID NIH HHS/United States
- T32 AI007313/AI/NIAID NIH HHS/United States
- F30 DA026310/DA/NIDA NIH HHS/United States
- T32 AI83196/AI/NIAID NIH HHS/United States
- P41 GM103426/GM/NIGMS NIH HHS/United States
- DP2 OD007237/OD/NIH HHS/United States
- Howard Hughes Medical Institute/United States
- P01 GM091743/GM/NIGMS NIH HHS/United States
- F31 AI116305/AI/NIAID NIH HHS/United States
- T32 GM008244/GM/NIGMS NIH HHS/United States
- R01 AI064046/AI/NIAID NIH HHS/United States
- DP2-OD007237/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

