Structural robustness of the gut mucosal microbiota is associated with Crohn's disease remission after surgery
- PMID: 26628508
- PMCID: PMC4893116
- DOI: 10.1136/gutjnl-2015-309184
Structural robustness of the gut mucosal microbiota is associated with Crohn's disease remission after surgery
Abstract
Objectives: Preventing postoperative recurrence after ileocolonic resection (ICR) for Crohn's disease (CD) is challenging. Defining the disturbances of the microbial composition and community structure after ICR and their link with early disease recurrence is crucial.
Design: Microbiota composition (fingerprinting and 16S rDNA sequencing) and community structure (correlation networks of bacterial species) were assessed from ileal mucosa sampled in 20 patients undergoing ICR and 6 months later during endoscopy from above (neoterminal ileum) and below (subanastomotic colon) the surgical anastomosis.
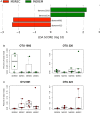
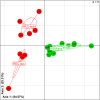
Results: ICR had a dramatic effect on gut microbial ecosystem. At surgery, CD mucosa harboured a dysbiotic microbiota with high proportions of α/β Proteobacteria and Bacilli. Six months later, half of the patients had recurrent lesions at ileocolonoscopy and presented higher numbers of Lachnospiraceae. Recurrence of endoscopic lesions was associated with enrichment in Enterococcus durans while patients in remission had increased proportions of Dorea longicatena and Bacteroides plebeius. Structural differences were striking between recurrence and remission microbiota; while the microbiota of patients with CD recurrence exhibited a loose community structure, the microbiota of patients in remission displayed communities that were robustly correlated to each other. Microbiota colonising the neoterminal ileum and subanastomotic colon 6 months after ICR only differed in patients with recurrence.
Conclusions: ICR modifies the gut microbiome. Remission after 6 months was associated with homogenous bacterial distribution around the anastomosis. Community structure and bacterial networks highlight target species, including Faecalibacterium prausnitzii and Ruminococcus gnavus, which may allow precise modulations of the overall microbial ecosystem towards remission pattern.
Keywords: BACTERIAL INTERACTIONS; INFLAMMATORY BOWEL DISEASE; INTESTINAL MICROBIOLOGY; MOLECULAR BIOLOGY; SURGICAL RESECTION.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/
Figures


Comment in
-
Microbiome-based companion diagnostics: no longer science fiction?Gut. 2016 Jun;65(6):896-7. doi: 10.1136/gutjnl-2015-311015. Epub 2016 Jan 22. Gut. 2016. PMID: 26801884 No abstract available.
-
The Mucosal Microbiome and Recurrence After Surgery for Crohn's Disease.Gastroenterology. 2016 Jun;150(7):1682-1684. doi: 10.1053/j.gastro.2016.04.026. Epub 2016 May 3. Gastroenterology. 2016. PMID: 27151260 No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
