Prediction of solution properties and dynamics of RNAs by means of Brownian dynamics simulation of coarse-grained models: Ribosomal 5S RNA and phenylalanine transfer RNA
- PMID: 26629336
- PMCID: PMC4666080
- DOI: 10.1186/s13628-015-0025-7
Prediction of solution properties and dynamics of RNAs by means of Brownian dynamics simulation of coarse-grained models: Ribosomal 5S RNA and phenylalanine transfer RNA
Abstract
Background: The possibility of validating biological macromolecules with locally disordered domains like RNA against solution properties is helpful to understand their function. In this work, we present a computational scheme for predicting global properties and mimicking the internal dynamics of RNA molecules in solution. A simple coarse-grained model with one bead per nucleotide and two types of intra-molecular interactions (elastic interactions and excluded volume interactions) is used to represent the RNA chain. The elastic interactions are modeled by a set of Hooke springs that form a minimalist elastic network. The Brownian dynamics technique is employed to simulate the time evolution of the RNA conformations.
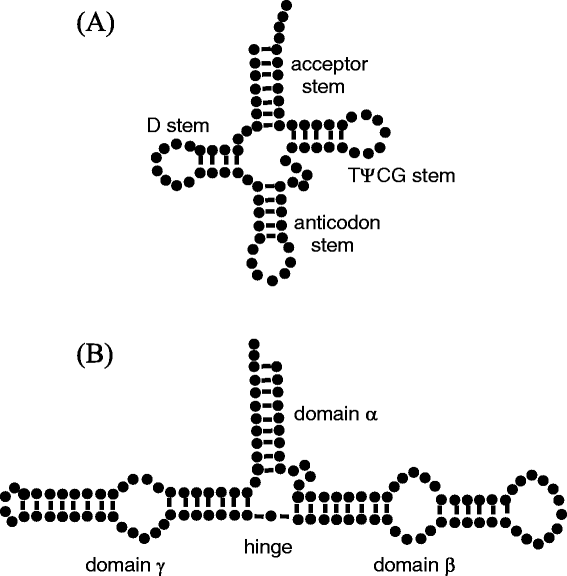
Results: That scheme is applied to the 5S ribosomal RNA of E. Coli and the yeast phenylalanine transfer RNA. From the Brownian trajectory, several solution properties (radius of gyration, translational diffusion coefficient, and a rotational relaxation time) are calculated. For the case of yeast phenylalanine transfer RNA, the time evolution and the probability distribution of the inter-arm angle is also computed.
Conclusions: The general good agreement between our results and some experimental data indicates that the model is able to capture the tertiary structure of RNA in solution. Our simulation results also compare quite well with other numerical data. An advantage of the scheme described here is the possibility of visualizing the real time macromolecular dynamics.
Keywords: Brownian dynamics; Coarse-grained model; Diffusion coefficients; Hydrodynamics; Internal dynamics; Ribosomal RNA; Transfer RNA.
Figures
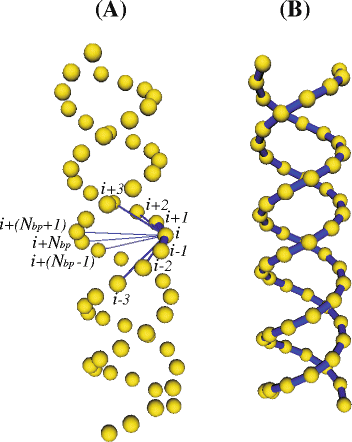


References
-
- Bloomfield VA, Crothers DM, Tinoco I. Nucleic Acids. Structures, Properties and Functions. Sausalito California: University Science Books; 2000.
LinkOut - more resources
Full Text Sources
Other Literature Sources

