Exploring the Gastrointestinal "Nemabiome": Deep Amplicon Sequencing to Quantify the Species Composition of Parasitic Nematode Communities
- PMID: 26630572
- PMCID: PMC4668017
- DOI: 10.1371/journal.pone.0143559
Exploring the Gastrointestinal "Nemabiome": Deep Amplicon Sequencing to Quantify the Species Composition of Parasitic Nematode Communities
Abstract
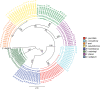
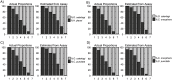
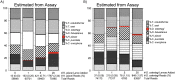
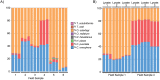
Parasitic helminth infections have a considerable impact on global human health as well as animal welfare and production. Although co-infection with multiple parasite species within a host is common, there is a dearth of tools with which to study the composition of these complex parasite communities. Helminth species vary in their pathogenicity, epidemiology and drug sensitivity and the interactions that occur between co-infecting species and their hosts are poorly understood. We describe the first application of deep amplicon sequencing to study parasitic nematode communities as well as introduce the concept of the gastro-intestinal "nemabiome". The approach is analogous to 16S rDNA deep sequencing used to explore microbial communities, but utilizes the nematode ITS-2 rDNA locus instead. Gastro-intestinal parasites of cattle were used to develop the concept, as this host has many well-defined gastro-intestinal nematode species that commonly occur as complex co-infections. Further, the availability of pure mono-parasite populations from experimentally infected cattle allowed us to prepare mock parasite communities to determine, and correct for, species representation biases in the sequence data. We demonstrate that, once these biases have been corrected, accurate relative quantitation of gastro-intestinal parasitic nematode communities in cattle fecal samples can be achieved. We have validated the accuracy of the method applied to field-samples by comparing the results of detailed morphological examination of L3 larvae populations with those of the sequencing assay. The results illustrate the insights that can be gained into the species composition of parasite communities, using grazing cattle in the mid-west USA as an example. However, both the technical approach and the concept of the 'nemabiome' have a wide range of potential applications in human and veterinary medicine. These include investigations of host-parasite and parasite-parasite interactions during co-infection, parasite epidemiology, parasite ecology and the response of parasite populations to both drug treatments and control programs.
Conflict of interest statement
Figures

References
-
- Stromberg BE, Gasbarre LC. Gastrointestinal nematode control programs with an emphasis on cattle. Vet Clin North Am Food Anim Pract 2006, November;22(3):543–65. - PubMed
-
- Parkins JJ, Holmes PH. Effects of gastrointestinal helminth parasites on ruminant nutrition. Nutrition Research Reviews 1989;2(01):227–46. - PubMed
-
- van Wyk JA, Cabaret J, Michael LM. Morphological identification of nematode larvae of small ruminants and cattle simplified. Vet Parasitol 2004, February 6;119(4):277–306. - PubMed
-
- Durette-Desset MC, Hugot JP, Darlu P, Chabaud AG. A cladistic analysis of the trichostrongyloidea (nematoda). Int J Parasitol 1999, July;29(7):1065–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

