Development of a fluorescent probe-based recombinase polymerase amplification assay for rapid detection of Orf virus
- PMID: 26631157
- PMCID: PMC4668657
- DOI: 10.1186/s12985-015-0440-z
Development of a fluorescent probe-based recombinase polymerase amplification assay for rapid detection of Orf virus
Abstract
Background: Orf virus (ORFV) is the causative agent of Orf (also known as contagious ecthyma or contagious papular dermatitis), a severe infectious skin disease in goats, sheep and other ruminants. The rapid detection of ORFV is of great importance in disease control and highly needed. A isothermal molecular diagnostic approach, termed recombinase polymerase amplification (RPA), is considered as an novel and rapid alternative techonology to PCR assay.
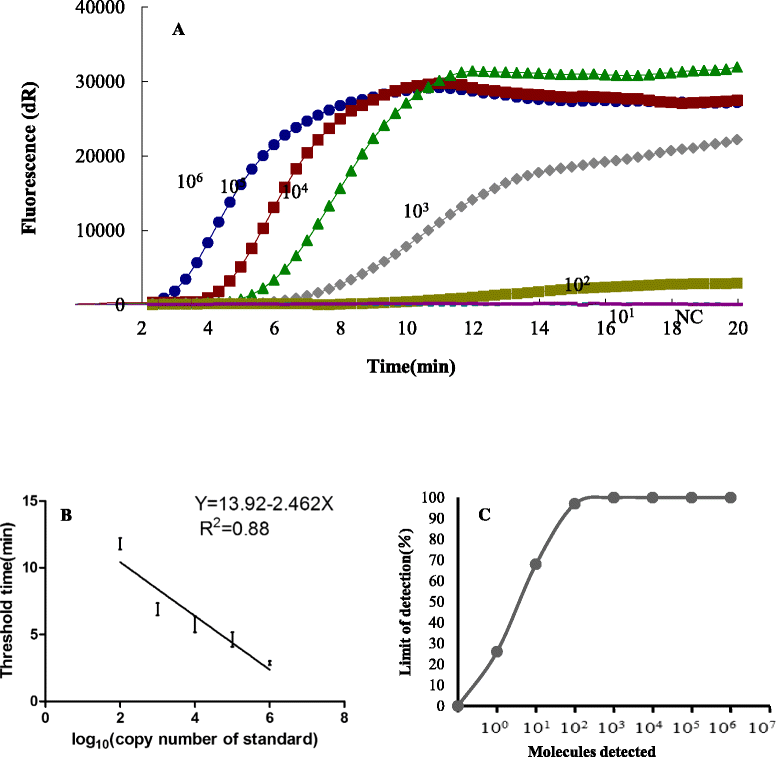
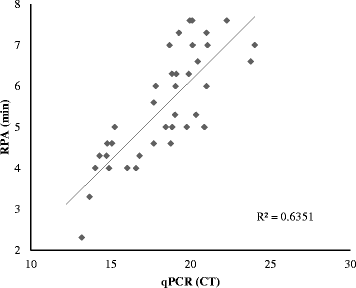
Results: In the present study, a novel fluorescent probe based on RPA assay (ORFV exo RPA assay) was developed. The developed ORFV exo RPA assay was capable of as low as 100 copies of ORFV DNA /reaction and was highly specific, with no cross-reaction with closely related viruses (capripox virus, foot-and-mouth disease virus or peste des petits ruminants virus). Further assessment with clinical samples showed that the developed ORFV exo RPA assay has good correlation with qPCR assays for detection of ORFV.
Conclusions: These results suggest that the developed ORFV exo RPA assay is suitable for rapid detection of ORFV.
Figures
Similar articles
-
Development of an isothermoal amplification-based assay for rapid visual detection of an Orf virus.Virol J. 2016 Mar 19;13:46. doi: 10.1186/s12985-016-0502-x. Virol J. 2016. PMID: 26993468 Free PMC article.
-
TaqMan based real-time duplex PCR for simultaneous detection and quantitation of capripox and orf virus genomes in clinical samples.J Virol Methods. 2014 Jun;201:44-50. doi: 10.1016/j.jviromet.2014.02.007. Epub 2014 Feb 16. J Virol Methods. 2014. PMID: 24552953
-
Development of a recombinase-aided amplification assay for detection of orf virus.J Virol Methods. 2020 Jun;280:113861. doi: 10.1016/j.jviromet.2020.113861. Epub 2020 Apr 25. J Virol Methods. 2020. PMID: 32343981
-
Recent advances in diagnostic approaches for orf virus.Appl Microbiol Biotechnol. 2023 Mar;107(5-6):1515-1523. doi: 10.1007/s00253-023-12412-8. Epub 2023 Feb 1. Appl Microbiol Biotechnol. 2023. PMID: 36723701 Review.
-
[Molecular characteristics and immune evasion strategies of ORFV: a review].Bing Du Xue Bao. 2012 May;28(3):278-84. Bing Du Xue Bao. 2012. PMID: 22764532 Review. Chinese.
Cited by
-
Rapid detection of highly pathogenic porcine reproductive and respiratory syndrome virus by a fluorescent probe-based isothermal recombinase polymerase amplification assay.Virus Genes. 2016 Dec;52(6):883-886. doi: 10.1007/s11262-016-1378-y. Epub 2016 Aug 17. Virus Genes. 2016. PMID: 27534870
-
Combined recombinase polymerase amplification/rkDNA-graphene oxide probing system for detection of SARS-CoV-2.Anal Chim Acta. 2021 May 8;1158:338390. doi: 10.1016/j.aca.2021.338390. Epub 2021 Mar 19. Anal Chim Acta. 2021. PMID: 33863409 Free PMC article.
-
Development of Isothermal Recombinase Polymerase Amplification Assay for Rapid Detection of Porcine Circovirus Type 2.Biomed Res Int. 2017;2017:8403642. doi: 10.1155/2017/8403642. Epub 2017 Mar 23. Biomed Res Int. 2017. PMID: 28424790 Free PMC article.
-
Development of real-time and lateral flow strip reverse transcription recombinase polymerase Amplification assays for rapid detection of peste des petits ruminants virus.Virol J. 2017 Feb 7;14(1):24. doi: 10.1186/s12985-017-0688-6. Virol J. 2017. PMID: 28173845 Free PMC article.
-
Rapid Onsite Visual Detection of Orf Virus Using a Recombinase-Aided Amplification Assay.Life (Basel). 2023 Feb 10;13(2):494. doi: 10.3390/life13020494. Life (Basel). 2023. PMID: 36836851 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

