The killing of macrophages by Corynebacterium ulcerans
- PMID: 26632348
- PMCID: PMC4871678
- DOI: 10.1080/21505594.2015.1125068
The killing of macrophages by Corynebacterium ulcerans
Abstract
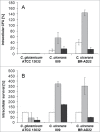
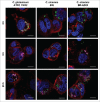
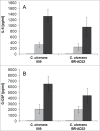
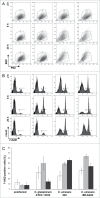
Corynebacterium ulcerans is an emerging pathogen transmitted by a zoonotic pathway with a very broad host spectrum to humans. Despite rising numbers of infections and potentially fatal outcomes, data on the molecular basis of pathogenicity are scarce. In this study, the interaction of 2 C. ulcerans isolates - one from an asymptomatic dog, one from a fatal case of human infection - with human macrophages was investigated. C. ulcerans strains were able to survive in macrophages for at least 20 hours. Uptake led to delay of phagolysosome maturation and detrimental effects on the macrophages as deduced from cytotoxicity measurements and FACS analyses. The data presented here indicate a high infectious potential of this emerging pathogen.
Keywords: Corynebacterium ulcerans; FACS; THP-1 cells; host-pathogen interaction; necrotic cell death.
Figures



References
-
- Tauch A, Sandbote J. The family Corynebacteriaceae In: Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F, eds. The Prokaryotes. Berlin Heidelberg: Springer; 2014:239-77
-
- Burkovski A. Cell envelope of corynebacteria: structure and influence on pathogenicity. ISRN Microbiol 2013; 2013:935736; PMID:23724339; http://dx.doi.org/10.1155/2013/935736 - DOI - PMC - PubMed
-
- Dorella FA, Pacheco LG, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res 2006; 37:201-18; PMID:16472520; http://dx.doi.org/10.1051/vetres:2005056 - DOI - PubMed
-
- Silva AS, Baraúna RA, de Sá PC, das Gracas DA, Carneiro AR, Thouvenin M, Azevedo V, Badell E, Guiso N, da Silva AL, Ramos RT. Draft genome sequence of Corynebacterium ulcerans FRC58, isolated from the bronchitic aspiration of a patient in France. Genome Announc 2014; 2:e01132-13; http://dx.doi.org/10.1128/genomeA.01132-13 - DOI - PMC - PubMed
-
- Gilbert R, Stewart FC. Corynebacterium ulcerans; a pathogenic microorganism resembling Corynebacterium diphtheriae. J Lab Clin Med 1927; 12:756-61
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
