The Novel Multiple Inner Primers-Loop-Mediated Isothermal Amplification (MIP-LAMP) for Rapid Detection and Differentiation of Listeria monocytogenes
- PMID: 26633345
- PMCID: PMC6332088
- DOI: 10.3390/molecules201219787
The Novel Multiple Inner Primers-Loop-Mediated Isothermal Amplification (MIP-LAMP) for Rapid Detection and Differentiation of Listeria monocytogenes
Abstract
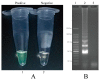
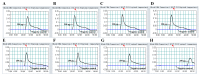
Here, a novel model of loop-mediated isothermal amplification (LAMP), termed multiple inner primers-LAMP (MIP-LAMP), was devised and successfully applied to detect Listeria monocytogenes. A set of 10 specific MIP-LAMP primers, which recognized 14 different regions of target gene, was designed to target a sequence in the hlyA gene. The MIP-LAMP assay efficiently amplified the target element within 35 min at 63 °C and was evaluated for sensitivity and specificity. The templates were specially amplified in the presence of the genomic DNA from L. monocytogenes. The limit of detection (LoD) of MIP-LAMP assay was 62.5 fg/reaction using purified L. monocytogenes DNA. The LoD for DNA isolated from serial dilutions of L. monocytogenes cells in buffer and in milk corresponded to 2.4 CFU and 24 CFU, respectively. The amplified products were analyzed by real-time monitoring of changes in turbidity, and visualized by adding Loop Fluorescent Detection Reagent (FD), or as a ladder-like banding pattern on gel electrophoresis. A total of 48 pork samples were investigated for L. monocytogenes by the novel MIP-LAMP method, and the diagnostic accuracy was shown to be 100% when compared to the culture-biotechnical method. In conclusion, the MIP-LAMP methodology was demonstrated to be a reliable, sensitive and specific tool for rapid detection of L. monocytogenes strains.
Keywords: HlyA gene amplification; L. monocytogenes detection; LAMP; LoD; MIP-LAMP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Rapid and sensitive detection of Listeria monocytogenes by loop-mediated isothermal amplification.Curr Microbiol. 2011 Dec;63(6):511-6. doi: 10.1007/s00284-011-0013-3. Epub 2011 Sep 21. Curr Microbiol. 2011. PMID: 21935669
-
Development of a loop-mediated isothermal amplification assay targeting lmo0753 gene for detection of Listeria monocytogenes in wastewater.Lett Appl Microbiol. 2019 Oct;69(4):264-270. doi: 10.1111/lam.13200. Epub 2019 Aug 9. Lett Appl Microbiol. 2019. PMID: 31323126
-
Reverse transcription - loop-mediated isothermal amplification assay for the rapid detection of pathogenic Listeria monocytogenes in meat products.Can J Microbiol. 2019 Dec;65(12):913-921. doi: 10.1139/cjm-2019-0114. Epub 2019 Sep 6. Can J Microbiol. 2019. PMID: 31491332
-
Lateral flow biosensor combined with loop-mediated isothermal amplification for simple, rapid, sensitive, and reliable detection of Brucella spp.Infect Drug Resist. 2019 Jul 30;12:2343-2353. doi: 10.2147/IDR.S211644. eCollection 2019. Infect Drug Resist. 2019. PMID: 31440069 Free PMC article. Review.
-
Antibody- and nucleic acid-based lateral flow immunoassay for Listeria monocytogenes detection.Anal Bioanal Chem. 2021 Jul;413(16):4161-4180. doi: 10.1007/s00216-021-03402-8. Epub 2021 May 26. Anal Bioanal Chem. 2021. PMID: 34041576 Review.
Cited by
-
Comparative seropositivity of Listeria monocytogenes in the serum of pregnant women with and without a history of abortion by serological and culture methods.SAGE Open Med. 2024 Jul 24;12:20503121241262189. doi: 10.1177/20503121241262189. eCollection 2024. SAGE Open Med. 2024. PMID: 39055278 Free PMC article.
-
Rapid and sensitive detection of Chlamydia trachomatis sexually transmitted infections in resource-constrained settings in Thailand at the point-of-care.PLoS Negl Trop Dis. 2018 Dec 20;12(12):e0006900. doi: 10.1371/journal.pntd.0006900. eCollection 2018 Dec. PLoS Negl Trop Dis. 2018. PMID: 30571705 Free PMC article. No abstract available.
-
Rapid and Sensitive Detection of Vibrio parahaemolyticus and Vibrio vulnificus by Multiple Endonuclease Restriction Real-Time Loop-Mediated Isothermal Amplification Technique.Molecules. 2016 Jan 19;21(1):E111. doi: 10.3390/molecules21010111. Molecules. 2016. PMID: 26797596 Free PMC article.
-
Enhancement of loop mediated isothermal amplification's sensitivity and speed by multiple inner primers for more efficient identification of Vibrio parahaemolyticus.MethodsX. 2023 Aug 21;11:102328. doi: 10.1016/j.mex.2023.102328. eCollection 2023 Dec. MethodsX. 2023. PMID: 37693654 Free PMC article.
-
Lab-Made Electronic Nose for Fast Detection of Listeria monocytogenes and Bacillus cereus.Vet Sci. 2020 Feb 9;7(1):20. doi: 10.3390/vetsci7010020. Vet Sci. 2020. PMID: 32050503 Free PMC article.
References
-
- Kathariou S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002;65:1811–1829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

