Metagenomic Analysis of Upwelling-Affected Brazilian Coastal Seawater Reveals Sequence Domains of Type I PKS and Modular NRPS
- PMID: 26633360
- PMCID: PMC4691048
- DOI: 10.3390/ijms161226101
Metagenomic Analysis of Upwelling-Affected Brazilian Coastal Seawater Reveals Sequence Domains of Type I PKS and Modular NRPS
Abstract
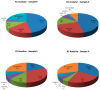
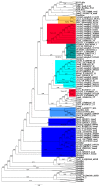
Marine environments harbor a wide range of microorganisms from the three domains of life. These microorganisms have great potential to enable discovery of new enzymes and bioactive compounds for industrial use. However, only ~1% of microorganisms from the environment can currently be identified through cultured isolates, limiting the discovery of new compounds. To overcome this limitation, a metagenomics approach has been widely adopted for biodiversity studies on samples from marine environments. In this study, we screened metagenomes in order to estimate the potential for new natural compound synthesis mediated by diversity in the Polyketide Synthase (PKS) and Nonribosomal Peptide Synthetase (NRPS) genes. The samples were collected from the Praia dos Anjos (Angel's Beach) surface water-Arraial do Cabo (Rio de Janeiro state, Brazil), an environment affected by upwelling. In order to evaluate the potential for screening natural products in Arraial do Cabo samples, we used KS (keto-synthase) and C (condensation) domains (from PKS and NRPS, respectively) to build Hidden Markov Models (HMM) models. From both samples, a total of 84 KS and 46 C novel domain sequences were obtained, showing the potential of this environment for the discovery of new genes of biotechnological interest. These domains were classified by phylogenetic analysis and this was the first study conducted to screen PKS and NRPS genes in an upwelling affected sample.
Keywords: NRPS; PKS; coastal environment; environmental genomics; metagenomics; upwelling.
Figures

Similar articles
-
[Screening and characterization of marine bacteria with antibacterial and cytotoxic activities, and existence of PKS I and NRPS genes in bioactive strains].Wei Sheng Wu Xue Bao. 2007 Apr;47(2):228-34. Wei Sheng Wu Xue Bao. 2007. PMID: 17552225 Chinese.
-
Discovery Strategies of Bioactive Compounds Synthesized by Nonribosomal Peptide Synthetases and Type-I Polyketide Synthases Derived from Marine Microbiomes.Mar Drugs. 2016 Apr 16;14(4):80. doi: 10.3390/md14040080. Mar Drugs. 2016. PMID: 27092515 Free PMC article. Review.
-
Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta.Appl Environ Microbiol. 2005 Aug;71(8):4840-9. doi: 10.1128/AEM.71.8.4840-4849.2005. Appl Environ Microbiol. 2005. PMID: 16085882 Free PMC article.
-
Phylogenetic analysis of type I polyketide synthase and nonribosomal peptide synthetase genes in Antarctic sediment.Extremophiles. 2008 Jan;12(1):97-105. doi: 10.1007/s00792-007-0107-9. Epub 2007 Aug 29. Extremophiles. 2008. PMID: 17726573
-
Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics.Biotechnol Lett. 2012 Aug;34(8):1393-403. doi: 10.1007/s10529-012-0919-2. Epub 2012 Apr 6. Biotechnol Lett. 2012. PMID: 22481301 Review.
Cited by
-
Mining the Yucatan Coastal Microbiome for the Identification of Non-Ribosomal Peptides Synthetase (NRPS) Genes.Toxins (Basel). 2020 May 26;12(6):349. doi: 10.3390/toxins12060349. Toxins (Basel). 2020. PMID: 32466531 Free PMC article.
-
Exploring the secrets of marine microorganisms: Unveiling secondary metabolites through metagenomics.Microb Biotechnol. 2024 Aug;17(8):e14533. doi: 10.1111/1751-7915.14533. Microb Biotechnol. 2024. PMID: 39075735 Free PMC article. Review.
-
Recovering Genomics Clusters of Secondary Metabolites from Lakes Using Genome-Resolved Metagenomics.Front Microbiol. 2018 Feb 20;9:251. doi: 10.3389/fmicb.2018.00251. eCollection 2018. Front Microbiol. 2018. PMID: 29515540 Free PMC article.
References
-
- Trindade-Silva A.E., Rua C., Silva G.G.Z., Dutilh B.E., Moreira A.P.B., Edwards R.A., Hajdu E., Lobo-Hajdu G., Vasconcelos A.T., Berlinck R.G., et al. Taxonomic and functional microbial signatures of the endemic marine sponge Arenosclera brasiliensis. PLoS ONE. 2012;7:e39905. doi: 10.1371/journal.pone.0039905. - DOI - PMC - PubMed
-
- Cury J.C., Araujo F.V., Coelho-Souza S.A., Peixoto R.S., Oliveira J.A.L., Santos H.F., Dávila A.M., Rosado A.S. Microbial diversity of a Brazilian coastal region influenced by an upwelling system and anthropogenic activity. PLoS ONE. 2011;6:e16553. doi: 10.1371/journal.pone.0016553. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

