Transcriptional program for nitrogen starvation-induced lipid accumulation in Chlamydomonas reinhardtii
- PMID: 26633994
- PMCID: PMC4667458
- DOI: 10.1186/s13068-015-0391-z
Transcriptional program for nitrogen starvation-induced lipid accumulation in Chlamydomonas reinhardtii
Abstract
Background: Algae accumulate lipids to endure different kinds of environmental stresses including macronutrient starvation. Although this response has been extensively studied, an in depth understanding of the transcriptional regulatory network (TRN) that controls the transition into lipid accumulation remains elusive. In this study, we used a systems biology approach to elucidate the transcriptional program that coordinates the nitrogen starvation-induced metabolic readjustments that drive lipid accumulation in Chlamydomonas reinhardtii.
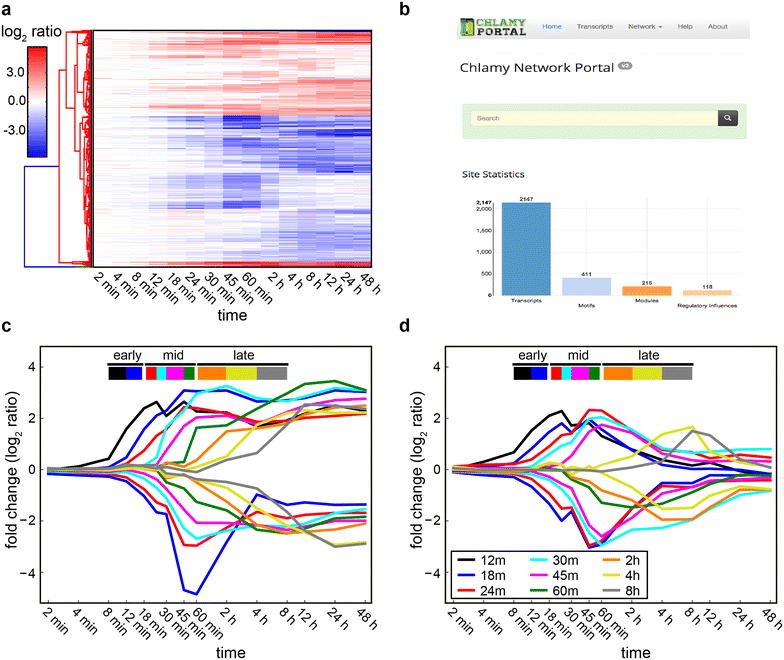
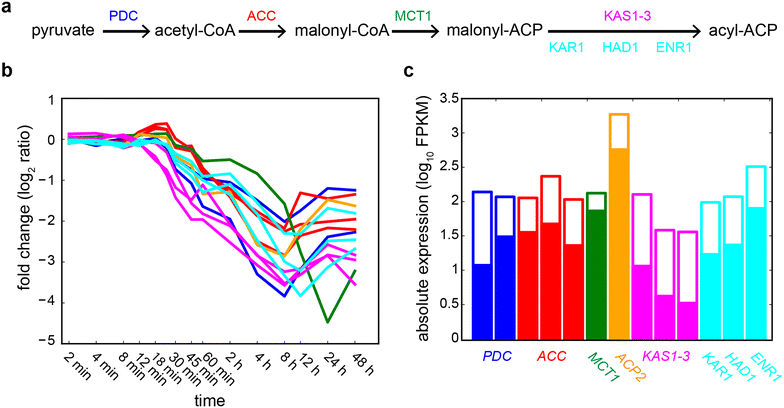
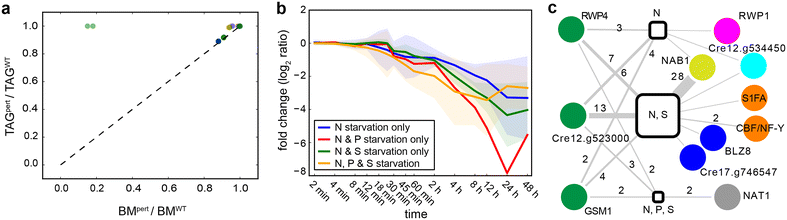
Results: We demonstrate that nitrogen starvation triggered differential regulation of 2147 transcripts, which were co-regulated in 215 distinct modules and temporally ordered as 31 transcriptional waves. An early-stage response was triggered within 12 min that initiated growth arrest through activation of key signaling pathways, while simultaneously preparing the intracellular environment for later stages by modulating transport processes and ubiquitin-mediated protein degradation. Subsequently, central metabolism and carbon fixation were remodeled to trigger the accumulation of triacylglycerols. Further analysis revealed that these waves of genome-wide transcriptional events were coordinated by a regulatory program orchestrated by at least 17 transcriptional regulators, many of which had not been previously implicated in this process. We demonstrate that the TRN coordinates transcriptional downregulation of 57 metabolic enzymes across a period of nearly 4 h to drive an increase in lipid content per unit biomass. Notably, this TRN appears to also drive lipid accumulation during sulfur starvation, while phosphorus starvation induces a different regulatory program. The TRN model described here is available as a community-wide web-resource at http://networks.systemsbiology.net/chlamy-portal.
Conclusions: In this work, we have uncovered a comprehensive mechanistic model of the TRN controlling the transition from N starvation to lipid accumulation. The program coordinates sequentially ordered transcriptional waves that simultaneously arrest growth and lead to lipid accumulation. This study has generated predictive tools that will aid in devising strategies for the rational manipulation of regulatory and metabolic networks for better biofuel and biomass production.
Keywords: Chlamydomonas reinhardtii; Lipid accumulation; Metabolic network; Network modeling; Phenotypic transition; Transcriptional regulatory network.
Figures

References
-
- Weers PMM, Gulati RD. Growth and reproduction of Daphnia galeata in response to changes in fatty acids, phosphorus, and nitrogen in Chlamydomonas reinhardtii. Limnol Oceanogr. 1997;42:1584–1589. doi: 10.4319/lo.1997.42.7.1584. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

