Systematic pan-cancer analysis of tumour purity
- PMID: 26634437
- PMCID: PMC4671203
- DOI: 10.1038/ncomms9971
Systematic pan-cancer analysis of tumour purity
Erratum in
-
Corrigendum: Systematic pan-cancer analysis of tumour purity.Nat Commun. 2016 Feb 5;7:10707. doi: 10.1038/ncomms10707. Nat Commun. 2016. PMID: 26848121 Free PMC article. No abstract available.
Abstract
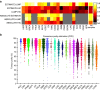
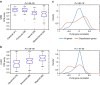
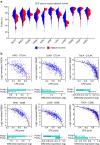
The tumour microenvironment is the non-cancerous cells present in and around a tumour, including mainly immune cells, but also fibroblasts and cells that comprise supporting blood vessels. These non-cancerous components of the tumour may play an important role in cancer biology. They also have a strong influence on the genomic analysis of tumour samples, and may alter the biological interpretation of results. Here we present a systematic analysis using different measurement modalities of tumour purity in >10,000 samples across 21 cancer types from the Cancer Genome Atlas. Patients are stratified according to clinical features in an attempt to detect clinical differences driven by purity levels. We demonstrate the confounding effect of tumour purity on correlating and clustering tumours with transcriptomics data. Finally, using a differential expression method that accounts for tumour purity, we find an immunotherapy gene signature in several cancer types that is not detected by traditional differential expression analyses.
Figures




References
-
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). - PubMed
-
- Junttila M. R. & de Sauvage F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013). - PubMed
-
- Schreiber R. D., Old L. J. & Smyth M. J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 331, 1565–1570 (2011). - PubMed
-
- Pages F. et al. Immune infiltration in human tumours: a prognostic factor that should not be ignored. Oncogene 29, 1093–1102 (2010). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

