An integrative analysis of small molecule transcriptional responses in the human malaria parasite Plasmodium falciparum
- PMID: 26637195
- PMCID: PMC4670519
- DOI: 10.1186/s12864-015-2165-1
An integrative analysis of small molecule transcriptional responses in the human malaria parasite Plasmodium falciparum
Abstract
Background: Transcriptional responses to small molecules can provide insights into drug mode of action (MOA). The capacity of the human malaria parasite, Plasmodium falciparum, to respond specifically to transcriptional perturbations has been unclear based on past approaches. Here, we present the most extensive profiling to date of the parasite's transcriptional responsiveness to thirty-one chemically and functionally diverse small molecules.
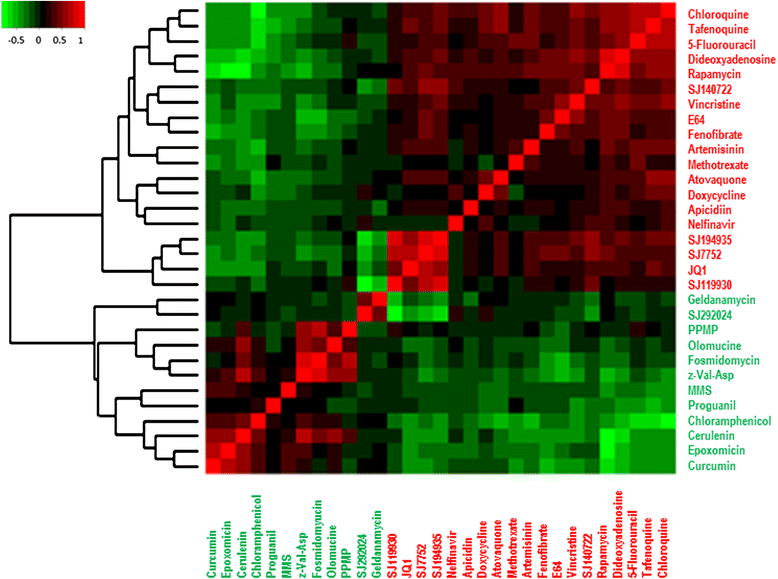
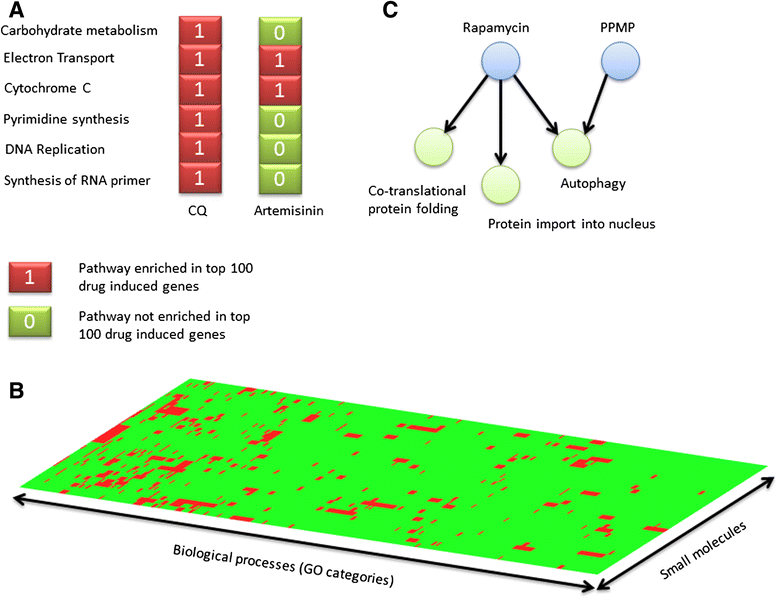
Methods: We exposed two laboratory strains of the human malaria parasite P. falciparum to brief treatments of thirty-one chemically and functionally diverse small molecules associated with biological effects across multiple pathways based on various levels of evidence. We investigated the impact of chemical composition and MOA on gene expression similarities that arise between perturbations by various compounds. To determine the target biological pathways for each small molecule, we developed a novel framework for encoding small molecule effects on a spectra of biological processes or GO functions that are enriched in the differentially expressed genes of a given small molecule perturbation.
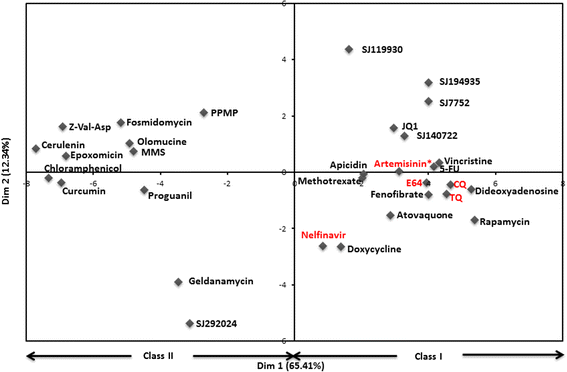
Results: We find that small molecules associated with similar transcriptional responses contain similar chemical features, and/ or have a shared MOA. The approach also revealed complex relationships between drugs and biological pathways that are missed by most exisiting approaches. For example, the approach was able to partition small molecule responses into drug-specific effects versus non-specific effects.
Conclusions: Our work provides a new framework for linking transcriptional responses to drug MOA in P. falciparum and can be generalized for the same purpose in other organisms.
Figures


References
-
- Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, et al. Artemisinin resistance: current status and scenarios for containment. Nat Rev Microbiol. 2010;8(4):272–280. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

