Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of β-Aryl Ether Bonds in Lignin
- PMID: 26637355
- PMCID: PMC4777856
- DOI: 10.1074/jbc.M115.694307
Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of β-Aryl Ether Bonds in Lignin
Abstract
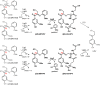
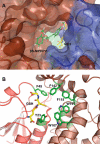
Lignin is a combinatorial polymer comprising monoaromatic units that are linked via covalent bonds. Although lignin is a potential source of valuable aromatic chemicals, its recalcitrance to chemical or biological digestion presents major obstacles to both the production of second-generation biofuels and the generation of valuable coproducts from lignin's monoaromatic units. Degradation of lignin has been relatively well characterized in fungi, but it is less well understood in bacteria. A catabolic pathway for the enzymatic breakdown of aromatic oligomers linked via β-aryl ether bonds typically found in lignin has been reported in the bacterium Sphingobium sp. SYK-6. Here, we present x-ray crystal structures and biochemical characterization of the glutathione-dependent β-etherases, LigE and LigF, from this pathway. The crystal structures show that both enzymes belong to the canonical two-domain fold and glutathione binding site architecture of the glutathione S-transferase family. Mutagenesis of the conserved active site serine in both LigE and LigF shows that, whereas the enzymatic activity is reduced, this amino acid side chain is not absolutely essential for catalysis. The results include descriptions of cofactor binding sites, substrate binding sites, and catalytic mechanisms. Because β-aryl ether bonds account for 50-70% of all interunit linkages in lignin, understanding the mechanism of enzymatic β-aryl ether cleavage has significant potential for informing ongoing studies on the valorization of lignin.
Keywords: X-ray crystallography; enzyme catalysis; enzyme mechanism; enzyme structure; lignin degradation; plant cell wall; protein structure; stereoselectivity; structural enzymology.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
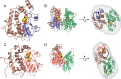


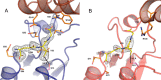

References
-
- Bugg T. D. H., Ahmad M., Hardiman E. M., and Rahmanpour R. (2011) Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 28, 1883–1896 - PubMed
-
- Bugg T. D. H., Ahmad M., Hardiman E. M., and Singh R. (2011) The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22, 394–400 - PubMed
-
- Leonowicz A., Cho N. S., Luterek J., Wilkolazka A., Wojtas-Wasilewska M., Matuszewska A., Hofrichter M., Wesenberg D., and Rogalski J. (2001) Fungal laccase: properties and activity on lignin. J. Basic Microbiol. 41, 185–227 - PubMed
-
- Martínez A. T., Speranza M., Ruiz-Dueñas F. J., Ferreira P., Camarero S., Guillén F., Martínez M. J., Gutiérrez A., and del Río J. C. (2005) Biodegradation of lignocellulosics: microbial chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 8, 195–204 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

