Munc13-4 Is a Rab11-binding Protein That Regulates Rab11-positive Vesicle Trafficking and Docking at the Plasma Membrane
- PMID: 26637356
- PMCID: PMC4751385
- DOI: 10.1074/jbc.M115.705871
Munc13-4 Is a Rab11-binding Protein That Regulates Rab11-positive Vesicle Trafficking and Docking at the Plasma Membrane
Abstract
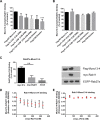
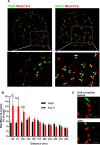
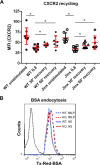
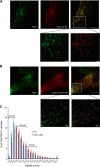
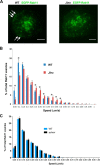
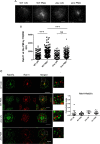
The small GTPase Rab11 and its effectors control trafficking of recycling endosomes, receptor replenishment and the up-regulation of adhesion and adaptor molecules at the plasma membrane. Despite recent advances in the understanding of Rab11-regulated mechanisms, the final steps mediating docking and fusion of Rab11-positive vesicles at the plasma membrane are not fully understood. Munc13-4 is a docking factor proposed to regulate fusion through interactions with SNAREs. In hematopoietic cells, including neutrophils, Munc13-4 regulates exocytosis in a Rab27a-dependent manner, but its possible regulation of other GTPases has not been explored in detail. Here, we show that Munc13-4 binds to Rab11 and regulates the trafficking of Rab11-containing vesicles. Using a novel Time-resolved Fluorescence Resonance Energy Transfer (TR-FRET) assay, we demonstrate that Munc13-4 binds to Rab11a but not to dominant negative Rab11a. Immunoprecipitation analysis confirmed the specificity of the interaction between Munc13-4 and Rab11, and super-resolution microscopy studies support the interaction of endogenous Munc13-4 with Rab11 at the single molecule level in neutrophils. Vesicular dynamic analysis shows the common spatio-temporal distribution of Munc13-4 and Rab11, while expression of a calcium binding-deficient mutant of Munc13-4 significantly affected Rab11 trafficking. Munc13-4-deficient neutrophils showed normal endocytosis, but the trafficking, up-regulation, and retention of Rab11-positive vesicles at the plasma membrane was significantly impaired. This correlated with deficient NADPH oxidase activation at the plasma membrane in response to Rab11 interference. Our data demonstrate that Munc13-4 is a Rab11-binding partner that regulates the final steps of Rab11-positive vesicle docking at the plasma membrane.
Keywords: Munc13-4; NADPH oxidase; Rab; Rab11; Rab27; docking; intracellular trafficking; neutrophil; recycling endosome; super-resolution microscopy.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Pfeffer S. R. (2001) Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11, 487–491 - PubMed
-
- Kelly E. E., Horgan C. P., and McCaffrey M. W. (2012) Rab11 proteins in health and disease. Biochem. Soc. Trans. 40, 1360–1367 - PubMed
-
- Takahashi S., Kubo K., Waguri S., Yabashi A., Shin H. W., Katoh Y., and Nakayama K. (2012) Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 125, 4049–4057 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

