Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2
- PMID: 26638071
- PMCID: PMC4671959
- DOI: 10.1016/j.cell.2015.11.005
Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2
Abstract
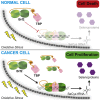
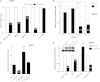
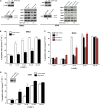
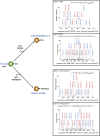
TFIIB-related factor 2 (Brf2) is a member of the family of TFIIB-like core transcription factors. Brf2 recruits RNA polymerase (Pol) III to type III gene-external promoters, including the U6 spliceosomal RNA and selenocysteine tRNA genes. Found only in vertebrates, Brf2 has been linked to tumorigenesis but the underlying mechanisms remain elusive. We have solved crystal structures of a human Brf2-TBP complex bound to natural promoters, obtaining a detailed view of the molecular interactions occurring at Brf2-dependent Pol III promoters and highlighting the general structural and functional conservation of human Pol II and Pol III pre-initiation complexes. Surprisingly, our structural and functional studies unravel a Brf2 redox-sensing module capable of specifically regulating Pol III transcriptional output in living cells. Furthermore, we establish Brf2 as a central redox-sensing transcription factor involved in the oxidative stress pathway and provide a mechanistic model for Brf2 genetic activation in lung and breast cancer.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures










References
-
- Biaglow J.E., Miller R.A. The thioredoxin reductase/thioredoxin system: novel redox targets for cancer therapy. Cancer Biol. Ther. 2005;4:6–13. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

